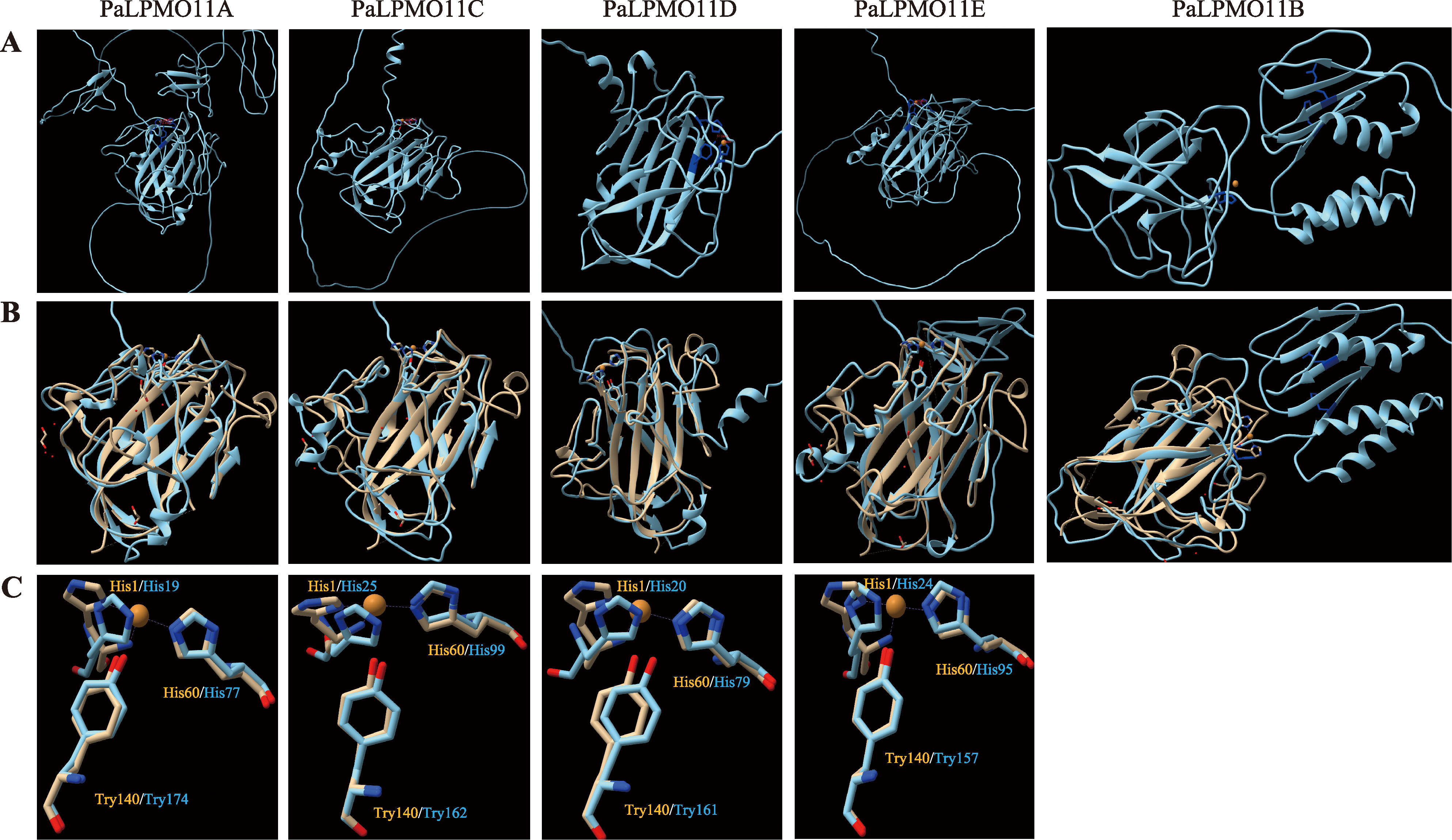
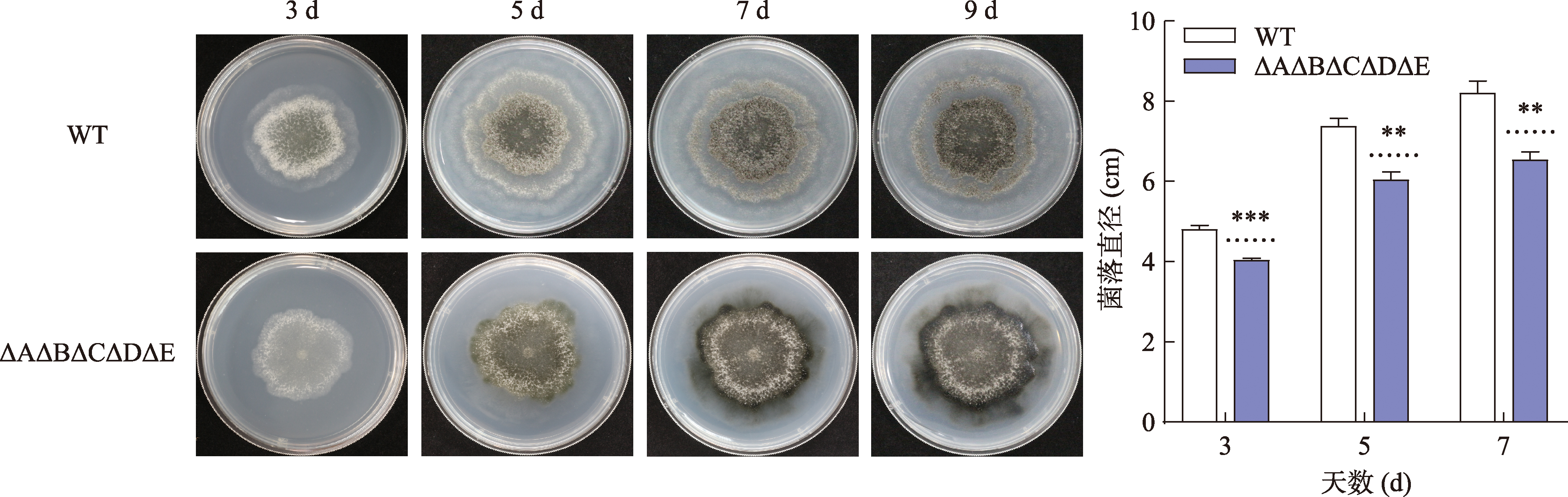
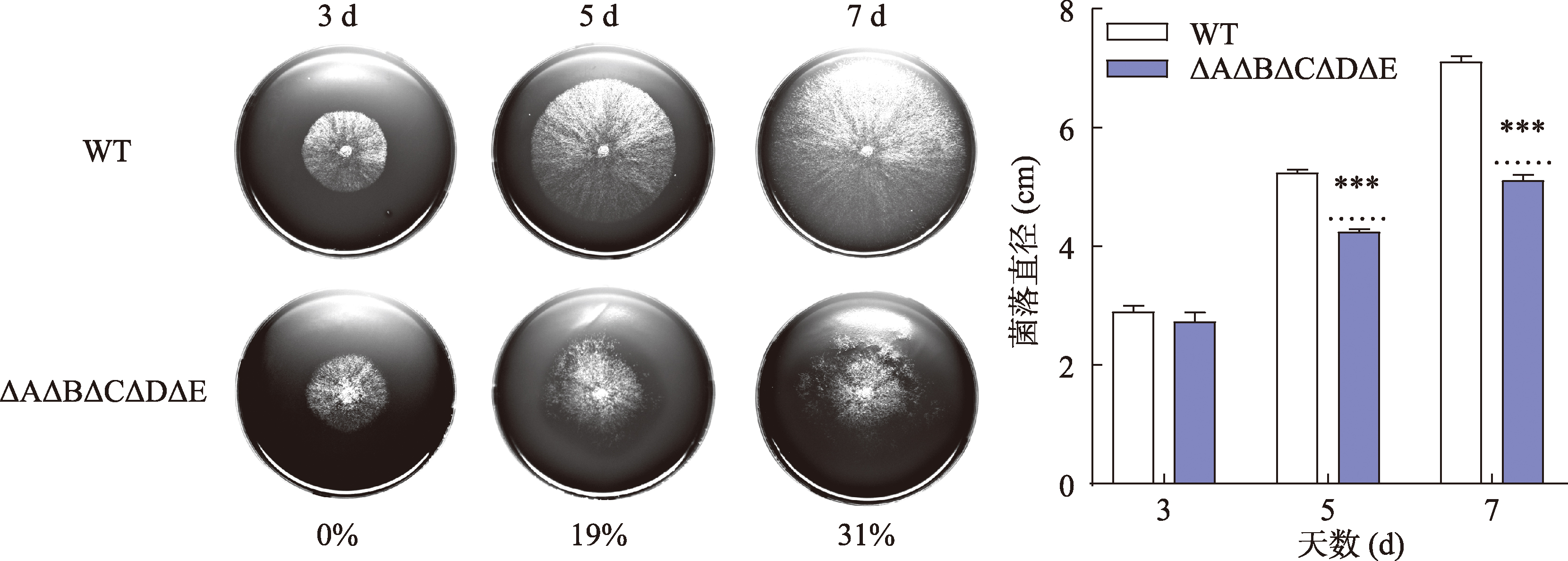
Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (12): 1128-1146.doi: 10.16288/j.yczz.23-223
• Research Article • Previous Articles Next Articles
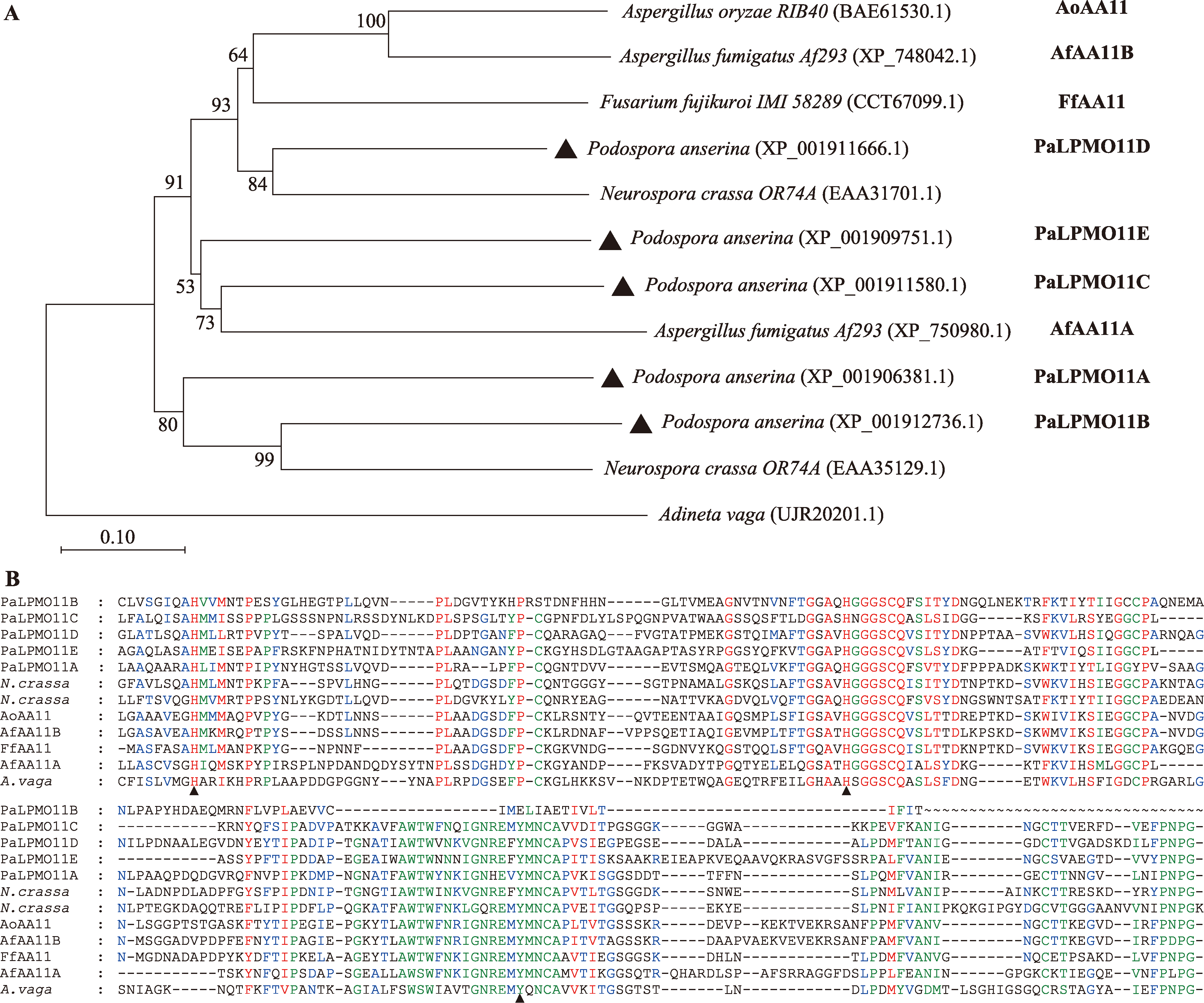
Identification and functional study of AA11 family polysaccharide monooxygenase genes in filamentous fungus Podospora anserina
Wenzhen Du1( ), Yuanjing Li2, Jialing Wu1, Siyu Chen1, Liang Jiang1, Gang Liu1, Ning Xie1(
), Yuanjing Li2, Jialing Wu1, Siyu Chen1, Liang Jiang1, Gang Liu1, Ning Xie1( )
)
- 1. Shenzhen Key Laboratory of Microbial Genetic Engineering, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen 518060, China
2. School of Materials and New Energy, South China Normal University, Shanwei 516622, China
-
Received:2023-08-21Revised:2023-10-28Online:2023-12-20Published:2023-11-10 -
Contact:Ning Xie E-mail:2100251038@email.szu.edu.cn;ning.xie@szu.edu.cn -
Supported by:National Key R&D Program of China(2021YFA0910800);Basic and Applied Basic Research Fund of Guangdong Province(2021A1515012166);Basic and Applied Basic Research Fund of Guangdong Province(2021A1515012118);National Natural Science Foundation of China(31601014);National Natural Science Foundation of China(22078199)
Cite this article
Wenzhen Du, Yuanjing Li, Jialing Wu, Siyu Chen, Liang Jiang, Gang Liu, Ning Xie. Identification and functional study of AA11 family polysaccharide monooxygenase genes in filamentous fungus Podospora anserina[J]. Hereditas(Beijing), 2023, 45(12): 1128-1146.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
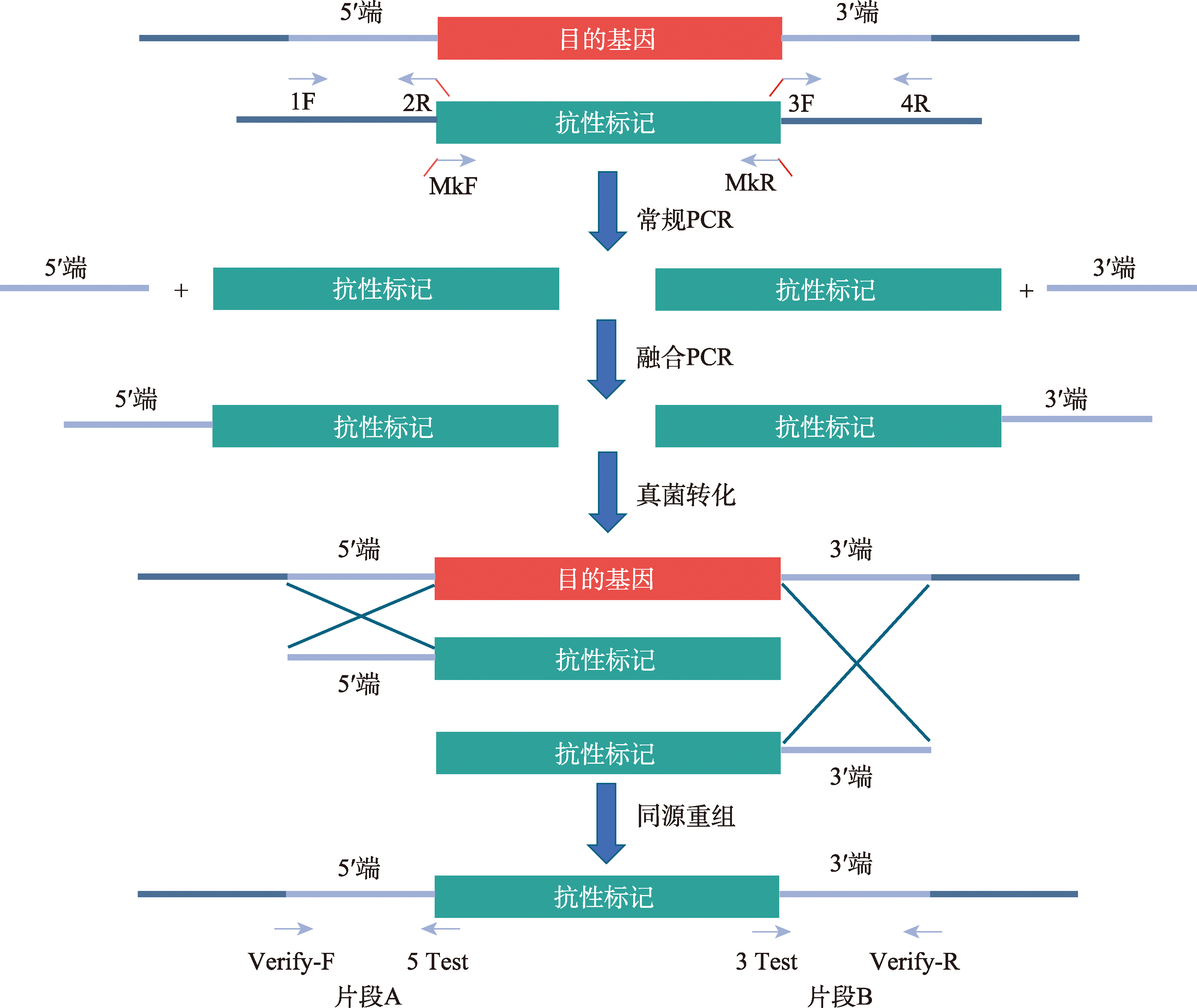
Table 1
Primers used in this study"
| 引物名称 | 序列 (5′→3′) | 用途 |
|---|---|---|
| PaLPMO11A_1F | CATTGACGCTCTCACCTTGG | 敲除PaLPMO11A基因 |
| PaLPMO11A_2R | CTATTTAACGACCCTGCCCTGAACCGATGCCCGACCTTCTGGTCCT | |
| PaLPMO11A_MKF | AGGACCAGAAGGTCGGGCATCGGTTCAGGGCAGGGTCGTTAAATAG | |
| PaLPMO11A_MKR | AGGTGACTACGATCCAGGCGCATCGAACTGGATCTCAACAGCGGTAAG | |
| PaLPMO11A_3F | CTTACCGCTGTTGAGATCCAGTTCGATGCGCCTGGATCGTAGTCACCT | |
| PaLPMO11A_4R | CCAGCCAACTTCCTCGTCAT | |
| PaLPMO11B_1F | CCCAGACAATGACCACACGAGT | 敲除PaLPMO11B基因 |
| PaLPMO11B_2R | CTATTTAACGACCCTGCCCTGAACCGCTGCTGCATGTCACTGAAGG | |
| PaLPMO11B_MkF | CCTTCAGTGACATGCAGCAGCGGTTCAGGGCAGGGTCGTTAAATAG | |
| PaLPMO11B_MkR | CAAGGGTAGCTGATGAGGCACATCGAACTGGATCTCAACAGCGGTAAG | |
| PaLPMO11B_3F | CTTACCGCTGTTGAGATCCAGTTCGATGTGCCTCATCAGCTACCCTTG | |
| PaLPMO11B_4R | CCGCGGTGAAGCTTGACACT | |
| PaLPMO11C_1F | ACAGGGATCATCGGTACAACTG | 敲除PaLPMO11C基因 |
| PaLPMO11C_2R | CTATTTAACGACCCTGCCCTGAACCGCTGGTTAGGTTCGAAGTCGAGT | |
| PaLPMO11C_MKF | ACTCGACTTCGAACCTAACCAGCGGTTCAGGGCAGGGTCGTTAAATAG | |
| PaLPMO11C_MKR | CTGGCTTCCAGTCGTCGATGCATCGAACTGGATCTCAACAGCGGTAAG | |
| PaLPMO11C_3F | CTTACCGCTGTTGAGATCCAGTTCGATGCATCGACGACTGGAAGCCAG | |
| PaLPMO11C_4R | CATCAGAACGACTCGCAGAT | |
| PaLPMO11D_1F | AGAACATCCAACGTGCGTGG | 敲除PaLPMO11D基因 |
| PaLPMO11D_2R | CTATTTAACGACCCTGCCCTGAACCGAATGACAGGACAAGTGCGGT | |
| PaLPMO11D_MKF | ACCGCACTTGTCCTGTCATTCGGTTCAGGGCAGGGTCGTTAAATAG | |
| PaLPMO11D_MKR | CCTGACAATGCTGAAACGACTGCATCGAACTGGATCTCAACAGCGGTAAG | |
| PaLPMO11D_3F | CTTACCGCTGTTGAGATCCAGTTCGATGCAGTCGTTTCAGCATTGTCAGG | |
| PaLPMO11D_4R | GCATCTCCATCAGCGCAGTG | |
| PaLPMO11E_1F | ACTAGAACGGCTCAGGCACACT | 敲除PaLPMO11E基因 |
| PaLPMO11E_2R | CTATTTAACGACCCTGCCCTGAACCGCGGATAGGCGAGTGATCGAT | |
| PaLPMO11E_MKF | ATCGATCACTCGCCTATCCGCGGTTCAGGGCAGGGTCGTTAAATAG | |
| PaLPMO11E_MKR | AGCTGCAGAAGTCCATCTCCCATCGAACTGGATCTCAACAGCGGTAAG | |
| PaLPMO11E_3F | CTTACCGCTGTTGAGATCCAGTTCGATGGGAGATGGACTTCTGCAGCT | |
| PaLPMO11E_4R | GCATGATATTCCCTCCACAGGT | |
| 5 Test | TGAGAAGCACACGGTCAC | 检测目的基因的敲除 |
| 3 Test | TCGGGGCGAAAACTCTC | |
| verify_PaLPMO11A_1F | AGGCCGGATACATACCGTTG | 验证PaLPMO11A基因敲除突变体 |
| verify_PaLPMO11A_2R | CACAATGTCCTCCAACACCG | |
| verify_PaLPMO11B_1F | ACCTTGCGAGAAGGTTGGTG | 验证PaLPMO11B基因敲除突变体 |
| verify_PaLPMO11B_2R | GAGGGAGGTGGCCTCTGAT | |
| verify_PaLPMO11C_1F | GAGCAAGTCCAGCCCTGACT | 验证PaLPMO11C基因敲除突变体 |
| verify_PaLPMO11C_2R | GCAGCGCAATCGTCGTGT | |
| verify_PaLPMO11D_1F | CAGCTGAGGCTAATGATCCG | 验证PaLPMO11D基因敲除突变体 |
| verify_PaLPMO11D_2R | CTCCTCGGCAGGTCACAAGT | |
| verify_PaLPMO11E_1F | AACCAGGTTCCCCAGGATCT | 验证PaLPMO11E基因敲除突变体 |
| verify_PaLPMO11E_2R | ATAACAGGCAGCTTGGCCTT |
| [1] |
Tran MH, Yu JH, Lee EY. Microwave-assisted two-step liquefaction of acetone-soluble lignin of silvergrass saccharification residue for production of biopolyol and biopolyurethane. Polymers (Basel), 2021, 13(9): 1491.
doi: 10.3390/polym13091491 |
| [2] |
Francois JM, Alkim C, Morin N. Engineering microbial pathways for production of bio-based chemicals from lignocellulosic sugars: current status and perspectives. Biotechnol Biofuels, 2020, 13: 118.
doi: 10.1186/s13068-020-01744-6 pmid: 32670405 |
| [3] |
Zhang ZR, Song JL, Han BX. Catalytic transformation of lignocellulose into chemicals and fuel products in ionic liquids. Chem Rev, 2017, 117(10): 6834-80.
doi: 10.1021/acs.chemrev.6b00457 pmid: 28535680 |
| [4] |
Upton BM, Kasko AM. Strategies for the conversion of lignin to high-value polymeric materials: review and perspective. Chem Rev, 2016, 116(4): 2275-306.
doi: 10.1021/acs.chemrev.5b00345 pmid: 26654678 |
| [5] |
Lyu LT, Chu YD, Zhang SF, Zhang Y, Huang QT, Wang S, Zhao ZK. Engineering the oleaginous yeast Rhodosporidium toruloides for improved resistance against inhibitors in biomass hydrolysates. Front Bioeng Biotechnol, 2021, 9: 768934.
doi: 10.3389/fbioe.2021.768934 |
| [6] |
Couturier M, Navarro D, Chevret D, Henrissat B, Piumi F, Ruiz-Dueñas FJ, Martinez AT, Grigoriev IV, Riley R, Lipzen A, Berrin JG, Master ER, Rosso MN. Enhanced degradation of softwood versus hardwood by the white-rot fungus Pycnoporus coccineus. Biotechnol Biofuels, 2015, 8: 216.
doi: 10.1186/s13068-015-0407-8 pmid: 26692083 |
| [7] |
Xia W, Xu XX, Qian LC, Shi PJ, Bai YG, Luo HY, Ma R, Yao B. Engineering a highly active thermophilic β-glucosidase to enhance its pH stability and saccharification performance. Biotechnol Biofuels, 2016, 9: 147.
doi: 10.1186/s13068-016-0560-8 pmid: 27446236 |
| [8] |
Calderaro F, Keser M, Akeroyd M, Bevers LE, Eijsink VGH, Várnai A, van den Berg MA. Characterization of an AA9 LPMO from Thielavia australiensis, TausLPMO9B, under industrially relevant lignocellulose saccharification conditions. Biotechnol Biofuels, 2020, 13(1): 195.
doi: 10.1186/s13068-020-01836-3 pmid: 33292403 |
| [9] |
Theibich YA, Sauer SPA, Leggio LL, Hedegård ED. Estimating the accuracy of calculated electron paramagnetic resonance hyperfine couplings for a lytic polysaccharide monooxygenase. Comput Struct Biotechnol J, 2021, 19: 555-67.
doi: 10.1016/j.csbj.2020.12.014 |
| [10] |
Span EA, Marletta MA. The framework of polysaccharide monooxygenase structure and chemistry. Curr Opin Struct Biol, 2015, 35: 93-99.
doi: 10.1016/j.sbi.2015.10.002 |
| [11] |
Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol, 2014, 10(2): 122-126.
doi: 10.1038/nchembio.1417 pmid: 24362702 |
| [12] |
Wang DM, Li J, Salazar-Alvarez G, McKee LS, Srivastava V, Sellberg JA, Bulone V, Hsieh YSY. Production of functionalised chitins assisted by fungal lytic polysaccharide monooxygenase. Green Chemistry, 2018, 20(9): 2091-2100.
doi: 10.1039/C8GC00422F |
| [13] |
Rieder L, Petrović D, Väljamäe P, Eijsink VGH, Sørlie M. Kinetic characterization of a putatively chitin-active LPMO reveals a preference for soluble substrates and absence of monooxygenase activity. ACS Catal, 2021, 11(18): 11685-11695.
doi: 10.1021/acscatal.1c03344 pmid: 34567832 |
| [14] |
Støpamo FG, Røhr ÅK, Mekasha S, Petrović DM, Várnai A, Eijsink VGH. Characterization of a lytic polysaccharide monooxygenase from Aspergillus fumigatus shows functional variation among family AA11 fungal LPMOs. J Biol Chem, 2021, 297(6): 101421.
doi: 10.1016/j.jbc.2021.101421 |
| [15] | Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol Biofuels, 2013, 6(1): 41. |
| [16] | Dicko M, Ferrari R, Tangthirasunun N, Gautier V, Lalanne C, Lamari F, Silar P. Lignin degradation and its use in signaling development by the coprophilous ascomycete Podospora anserina. J Fungi (Basel), 2020, 6(4): 278. |
| [17] |
El-Khoury R, Sellem CH, Coppin E, Boivin A, Maas MFPM, Debuchy R, Sainsard-Chanet A. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr Genet, 2008, 53(4): 249-258.
doi: 10.1007/s00294-008-0180-3 pmid: 18265986 |
| [18] |
Goswami RS. Targeted gene replacement in fungi using a split-marker approach. Methods Mol Biol, 2012, 835: 255-269.
doi: 10.1007/978-1-61779-501-5_16 pmid: 22183659 |
| [19] | Yan LY, Zhang HJ, Zheng YQ, Cong YQ, Liu CT, Fan F, Zheng C, Yuan GL, Pan G, Yuan DY, Duan MJ. Transcription factor OsMADS25 improves rice tolerance to cold stress. Hereditas (Beijing), 2021, 43(11): 1078-1087. |
| 闫凌月, 张豪健, 郑雨晴, 丛韫起, 刘次桃, 樊帆, 郑铖, 袁贵龙, 潘根, 袁定阳, 段美娟. 转录因子OsMADS25提高水稻对低温的耐受性. 遗传, 2021, 43(11): 1078-1087. | |
| [20] | Tangthirasunun N, Navarro D, Garajova S, Chevret D, Tong LCH, Gautier V, Hyde KD, Silar P, Berrin JG. Inactivation of cellobiose dehydrogenases modifies the cellulose degradation mechanism of Podospora anserina. Appl Environ Microbiol, 2017, 83(2): e02716-16. |
| [21] |
Dashtban M, Maki M, Leung KT, Mao CQ, Qin WS. Cellulase activities in biomass conversion: measurement methods and comparison. Crit Rev Biotechnol, 2010, 30(4): 302-309.
doi: 10.3109/07388551.2010.490938 pmid: 20868219 |
| [22] |
Thankappan S, Kandasamy S, Joshi B, Sorokina KN, Taran OP, Uthandi S. Bioprospecting thermophilic glycosyl hydrolases, from hot springs of Himachal Pradesh, for biomass valorization. AMB Express, 2018, 8(1): 168.
doi: 10.1186/s13568-018-0690-4 pmid: 30324223 |
| [23] |
Bennati-Granier C, Garajova S, Champion C, Grisel S, Haon M, Zhou SM, Fanuel M, Ropartz D, Rogniaux H, Gimbert I, Record E, Berrin JG.Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnol Biofuels, 2015, 8: 90.
doi: 10.1186/s13068-015-0274-3 pmid: 26136828 |
| [24] | Demoor A, Silar P, Brun S. Appressorium: the breakthrough in Dikarya. J Fungi (Basel), 2019, 5(3): 72. |
| [25] |
Xie N, Ruprich-Robert G, Silar P, Chapeland-Leclerc F. Bilirubin oxidase-like proteins from Podospora anserina: promising thermostable enzymes for application in transformation of plant biomass. Environ Microbiol, 2015, 17(3): 866-875.
doi: 10.1111/emi.2015.17.issue-3 |
| [26] |
Fu C, Thielhelm TP, Heitman J. Unisexual reproduction promotes competition for mating partners in the global human fungal pathogen Cryptococcus deneoformans. PLoS Genet, 2019, 15(9): e1008394.
doi: 10.1371/journal.pgen.1008394 |
| [27] |
Wang M, Gu BL, Huang J, Jiang S, Chen YJ, Yin YL, Pan YF, Yu GJ, Li YM, Wong BHC, Liang Y, Sun H. Transcriptome and proteome exploration to provide a resource for the study of Agrocybe aegerita. PLoS One, 2013, 8(2): e56686.
doi: 10.1371/journal.pone.0056686 |
| [28] |
Orban A, Weber A, Herzog R, Hennicke F, Rühl M. Transcriptome of different fruiting stages in the cultivated mushroom Cyclocybe aegerita suggests a complex regulation of fruiting and reveals enzymes putatively involved in fungal oxylipin biosynthesis. BMC Genomics, 2021, 22(1): 324.
doi: 10.1186/s12864-021-07648-5 pmid: 33947322 |
| [29] |
Schumacher DI, Lütkenhaus R, Altegoer F, Teichert I, Kück U, Nowrousian M.The transcription factor PRO44 and the histone chaperone ASF1 regulate distinct aspects of multicellular development in the filamentous fungus Sordaria macrospora. BMC Genet, 2018, 19(1): 112.
doi: 10.1186/s12863-018-0702-z pmid: 30545291 |
| [30] |
Ismail HF, Hashim Z, Soon WT, Rahman NSA, Zainudin AN, Majid FAA. Comparative study of herbal plants on the phenolic and flavonoid content, antioxidant activities and toxicity on cells and zebrafish embryo. J Tradit Complement Med, 2017, 7(4): 452-465.
doi: 10.1016/j.jtcme.2016.12.006 pmid: 29034193 |
| [31] |
Zhang HR, Li BX, Sun ZM, Zhou H, Zhang SS. Integration of intracellular telomerase monitoring by electrochemiluminescence technology and targeted cancer therapy by reactive oxygen species. Chem Sci, 2017, 8(12): 8025-8029.
doi: 10.1039/c7sc03772d pmid: 29568450 |
| [32] |
Kalyani D, Tiwari MK, Li JL, Kim SC, Kalia VC, Kang YC, Lee JK. A highly efficient recombinant laccase from the yeast Yarrowia lipolytica and its application in the hydrolysis of biomass. PLoS One, 2015, 10(3): e0120156.
doi: 10.1371/journal.pone.0120156 |
| [33] |
Abraham RE, Wong CS, Puri M. Enrichment of cellulosic waste hemp (Cannabis sativa) hurd into non-toxic microfibres. Materials (Basel), 2016, 9(7): 562.
doi: 10.3390/ma9070562 |
| [34] |
Zhang XJ, Qu YB, Qin YQ.Expression and chromatin structures of cellulolytic enzyme gene regulated by heterochromatin protein 1. Biotechnol Biofuels, 2016, 9: 206.
pmid: 27729944 |
| [35] |
Laurent CVFP, Sun PC, Scheiblbrandner S, Csarman F, Cannazza P, Frommhagen M, Oostenbrink C, Kabel MA, Ludwig R. Influence of lytic polysaccharide monooxygenase active site segments on activity and affinity. Int J Mol Sci, 2019, 20(24): 6219.
doi: 10.3390/ijms20246219 |
| [36] | Silar P. Podospora anserina: from laboratory to biotechnology. Genomics of soil- and plant-associated fungi. 2013, 283-309. |
| [37] |
Bernhardt D, Hamann A, Osiewacz HD. The role of mitochondria in fungal aging. Curr Opin Microbiol, 2014, 22: 1-7.
doi: 10.1016/j.mib.2014.09.007 pmid: 25299751 |
| [38] |
Pellavio G, Sommi P, Anselmi-Tamburini U, DeMichelis MP, Coniglio S, Laforenza U. Cerium oxide nanoparticles regulate oxidative stress in hela cells by increasing the aquaporin-mediated hydrogen peroxide permeability. Int J Mol Sci, 2022, 23(18): 10837.
doi: 10.3390/ijms231810837 |
| [39] | Gao H, Liu XP, Tian KM, Meng YC, Yu CC, Peng YF. Insight into the protective effect of salidroside against H2O2-induced injury in H9C2 cells. Oxid Med Cell Longev, 2021, 2021: 1060271. |
| [40] |
Wiemer M, Osiewacz HD. Effect of paraquat-induced oxidative stress on gene expression and aging of the filamentous ascomycete Podospora anserina. Microb Cell, 2014, 1(7): 225-240.
doi: 10.15698/mic2014.07.155 pmid: 28357247 |
| [41] | Wu LW, Ren DY, Hu SK, Li GM, Dong GJ, Jiang L, Hu XM, Ye WJ, Cui YT, Zhu L, Hu J, Zhang GH, Gao ZY, Zeng DL, Qian Q, Guo LB. Down-regulation of a nicotinate phosphoribosyltransferase gene, OsNaPRT1, leads to withered leaf tips. Plant Physiol, 2016, 171(2): 1085-1098. |
| [42] |
Bian HY, Wu XX, Luo J, Qiao YZ, Fang GG, Dai HQ. Valorization of alkaline peroxide mechanical pulp by metal chloride-assisted hydrotropic pretreatment for enzymatic saccharification and cellulose nanofibrillation. Polymers (Basel), 2019, 11(2): 331.
doi: 10.3390/polym11020331 |
| [43] |
Couturier M, Tangthirasunun N, Ning X, Brun S, Gautier V, Bennati-Granier C, Silar P, Berrin JG. Plant biomass degrading ability of the coprophilic ascomycete fungus Podospora anserina. Biotechnol Adv, 2016, 34(5): 976-983.
doi: S0734-9750(16)30067-2 pmid: 27263000 |
| [44] |
Brun S, Malagnac F, Bidard F, Lalucque H, Silar P. Functions and regulation of the Nox family in the filamentous fungus Podospora anserina: a new role in cellulose degradation. Mol Microbiol, 2009, 74(2): 480-496.
doi: 10.1111/mmi.2009.74.issue-2 |
| [45] |
Xie N, Chapeland-Leclerc F, Silar P, Ruprich-Robert G. Systematic gene deletions evidences that laccases are involved in several stages of wood degradation in the filamentous fungus Podospora anserina. Environ Microbiol, 2014, 16(1): 141-161.
doi: 10.1111/1462-2920.12253 pmid: 24102726 |
| [1] | Lan Wang, Fan Zeng, Rongfeng Huang, Shu Lin, Zhihui Zhang, Min-Dian Li. Adipocyte reconstitution of Npy4r gene in Npy4r silenced mice promotes diet-induced obesity [J]. Hereditas(Beijing), 2023, 45(2): 144-155. |
| [2] | Meizhen Liu, Liren Wang, Yongmei Li, Xueyun Ma, Honghui Han, Dali Li. Generation of genetically modified rat models via the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2023, 45(1): 78-87. |
| [3] | Minting Lin, Lulu Lai, Miao Zhao, Biwei Lin, Xiangping Yao. Construction of a striatum-specific Slc20a2 gene knockout mice model by CRISPR/Cas9 AAV system [J]. Hereditas(Beijing), 2020, 42(10): 1017-1027. |
| [4] | Junbo Tang, Haowei Cao, Rui Xu, Dandan Zhang, Juan Huang. Mutant generation of the testis genes and phenotype analyses in Drosophila [J]. Hereditas(Beijing), 2018, 40(6): 478-487. |
| [5] | Yuanyuan Pan, Gang Liu. Research advances on molecular regulation of filamentous fungal secondary metabolism in China [J]. Hereditas(Beijing), 2018, 40(10): 874-887. |
| [6] | Honghua Li,Gang Liu. The application of CRISPR/Cas9 in genome editing of filamentous fungi [J]. Hereditas(Beijing), 2017, 39(5): 355-367. |
| [7] | Fenghua Zhang, Houpeng Wang, Siyu Huang, Feng Xiong, Zuoyan Zhu, Yonghua Sun. A comparison of the knockout efficiencies of two codon-optimized Cas9 coding sequences in zebrafish embryos [J]. HEREDITAS(Beijing), 2016, 38(2): 144-154. |
| [8] | Xiaoli Wang,Chuang Jiang,Jianhua Liu,Xipeng Liu. An efficient genetic knockout system based on linear DNA fragment homologous recombination for halophilic archaea [J]. HEREDITAS(Beijing), 2015, 37(4): 388-395. |
| [9] | Hui Wang, Guang Li, Yiquan Wang. Generating amphioxus Hedgehog knockout mutants and phenotype analysis [J]. HEREDITAS(Beijing), 2015, 37(10): 1036-1043. |
| [10] | Feida Li, Yong Li, Huan Liu, Huanhuan Zhang, Chuxin Liu, Xingju Zhang, Hongwei Dou, Wenxian Yang, Yutao Du. Production of GHR double-allelic knockout Bama pig by TALENs and handmade cloning [J]. HEREDITAS(Beijing), 2014, 36(9): 903-911. |
| [11] | CAO Sui-Zhong YUE Cheng-He LI Xi-Rui FENG Chong LONG Chuan PAN Deng-Ke. Production of myostatin gene knockout Wuzhishan miniature pig fibroblasts with zinc-finger nucleases [J]. HEREDITAS, 2013, 35(6): 778-785. |
| [12] | CHEN Xian-Zhong, CHEN Wei, FAN Liu, WANG Zheng-Xiang. Genomics and metabolic engineering of filamentous fungi in the post-genomics era [J]. HEREDITAS, 2011, 33(10): 1067-1078. |
| [13] | WANG Jiao-Yu, DU Xin-Fa, CHAI Rong-Yao, SUN Guo-Chang, LIN Fu-Cheng. Strategies of targeted gene replacement in filamentous fungi [J]. HEREDITAS, 2007, 29(7): 898-904. |
| [14] | TENG Yan, YANG Xiao. Gene targeting: the beginning of a new era in genetics [J]. HEREDITAS, 2007, 29(11): 1291-1298. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||