Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (8): 640-648.doi: 10.16288/j.yczz.24-068
• Research Article • Previous Articles Next Articles
Application effectiveness of “Youxin-1” in genetic diversity and structure analysis of local chickens
Mengyu Wang1,2( ), Chenghao Zhou2(
), Chenghao Zhou2( ), Qian Xue2,3, Jianmei Yin2, Yixiu Jiang2, Huiyong Zhang2, Guohui Li2, Wei Han1,2(
), Qian Xue2,3, Jianmei Yin2, Yixiu Jiang2, Huiyong Zhang2, Guohui Li2, Wei Han1,2( )
)
- 1. Poultry Institute, Chinese Academy of Agricultural Sciences, Yangzhou 225125, China
2. Poultry Institute of Jiangsu Province, Yangzhou 225125, China
3. Science and Technology Innovation Co., Ltd., Poultry Institute of JiangsuProvince, Yangzhou 225125, China
-
Received:2024-03-14Revised:2024-06-12Online:2024-08-20Published:2024-06-24 -
Contact:Wei Han E-mail:2664935435@qq.com;604242123@qq.com;hanwei830@163.com -
Supported by:National Key Research and Development Program of China(2021YFD1200302);Seed Industry Revitalization Project of Jiangsu Province(JBGS[2021]029);Natural Science Foundation Project of Jiangsu Province(BK20221285)
Cite this article
Mengyu Wang, Chenghao Zhou, Qian Xue, Jianmei Yin, Yixiu Jiang, Huiyong Zhang, Guohui Li, Wei Han. Application effectiveness of “Youxin-1” in genetic diversity and structure analysis of local chickens[J]. Hereditas(Beijing), 2024, 46(8): 640-648.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
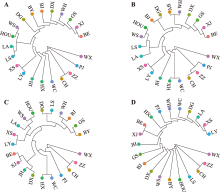
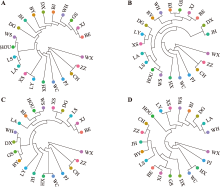
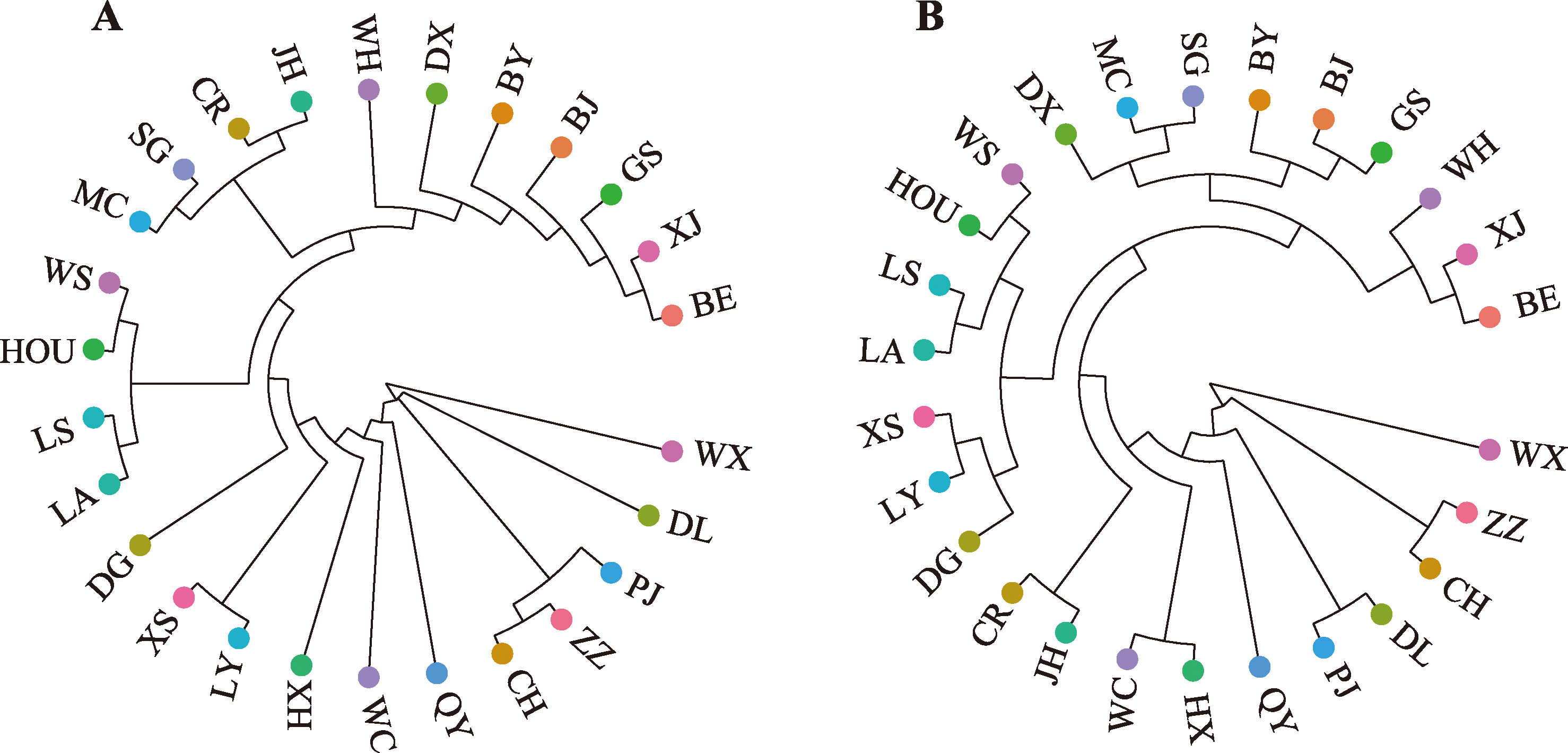
Table 1
Genetic diversity statistics calculated based on RADseq"
| 品种 | 观察杂合度(Ho) | 多态信息含量(PIC) | 近交系数 (FROH) | 平均遗传分化系数 (Fst) |
|---|---|---|---|---|
| 白耳黄鸡(BE) | 0.2049 | 0.2712 | 0.1562 | 0.1632 |
| 边鸡(BJ) | 0.1833 | 0.2717 | 0.1697 | 0.1443 |
| 北京油鸡(BY) | 0.1887 | 0.2586 | 0.2328 | 0.1781 |
| 茶花鸡(CH) | 0.2104 | 0.2844 | 0.1547 | 0.1592 |
| 大骨鸡(DG) | 0.2121 | 0.2953 | 0.1373 | 0.1362 |
| 东乡绿壳蛋鸡(DX) | 0.1876 | 0.262 | 0.1806 | 0.1614 |
| 固始鸡(GS) | 0.1901 | 0.2558 | 0.2277 | 0.1715 |
| 天津猴鸡(HOU) | 0.2214 | 0.2822 | 0.2077 | 0.1785 |
| 惠阳胡须鸡(HX) | 0.2179 | 0.3016 | 0.0972 | 0.1235 |
| 金湖乌凤鸡(JH) | 0.2039 | 0.284 | 0.169 | 0.1423 |
| 琅琊鸡(LA) | 0.2535 | 0.3182 | 0.0536 | 0.1028 |
| 狼山鸡(LS) | 0.205 | 0.2691 | 0.1989 | 0.1795 |
| 鹿苑鸡(LY) | 0.2447 | 0.3202 | 0.048 | 0.1026 |
| 瓢鸡(PJ) | 0.2359 | 0.3149 | 0.0181 | 0.1079 |
| 文昌鸡(WC) | 0.224 | 0.2981 | 0.0732 | 0.1277 |
| 安义瓦灰鸡(WH) | 0.2172 | 0.2879 | 0.1369 | 0.1408 |
| 汶上芦花鸡(WS) | 0.2163 | 0.2853 | 0.1703 | 0.1515 |
| 大围山微型鸡(WX) | 0.2154 | 0.2886 | 0.1264 | 0.1455 |
| 仙居鸡(XJ) | 0.2047 | 0.2816 | 0.1445 | 0.1503 |
| 萧山鸡(XS) | 0.2146 | 0.2842 | 0.1657 | 0.1544 |
| 藏鸡(ZZ) | 0.2127 | 0.2979 | 0.145 | 0.1396 |
Table 2
Genetic diversity statistics calculated based on 23K chips"
| 品种 | 观察杂合度(Ho) | 多态信息含量(PIC) | 近交系数 (FROH) | 平均遗传分化系数 (Fst) |
|---|---|---|---|---|
| 白耳黄鸡(BE) | 0.1955 | 0.2714 | 0.1543 | 0.1902 |
| 边鸡(BJ) | 0.1856 | 0.2813 | 0.1787 | 0.1598 |
| 北京油鸡(BY) | 0.1735 | 0.2482 | 0.1911 | 0.2001 |
| 茶花鸡(CH) | 0.2066 | 0.2867 | 0.1444 | 0.1927 |
| 大骨鸡(DG) | 0.2205 | 0.2963 | 0.1364 | 0.1557 |
| 东乡绿壳蛋鸡(DX) | 0.1411 | 0.2413 | 0.2147 | 0.1811 |
| 固始鸡(GS) | 0.1824 | 0.2577 | 0.1899 | 0.1862 |
| 天津猴鸡(HOU) | 0.1966 | 0.2644 | 0.2105 | 0.2010 |
| 惠阳胡须鸡(HX) | 0.2096 | 0.3085 | 0.1201 | 0.1438 |
| 金湖乌凤鸡(JH) | 0.2008 | 0.3016 | 0.1724 | 0.1718 |
| 琅琊鸡(LA) | 0.2472 | 0.3156 | 0.0736 | 0.1273 |
| 狼山鸡(LS) | 0.1867 | 0.2556 | 0.1609 | 0.2039 |
| 鹿苑鸡(LY) | 0.213 | 0.3102 | 0.0568 | 0.1229 |
| 瓢鸡(PJ) | 0.2242 | 0.3139 | 0.0780 | 0.1282 |
| 文昌鸡(WC) | 0.2493 | 0.3174 | 0.0940 | 0.1453 |
| 安义瓦灰鸡(WH) | 0.2135 | 0.2888 | 0.1331 | 0.1615 |
| 汶上芦花鸡(WS) | 0.2165 | 0.2862 | 0.1657 | 0.1729 |
| 大围山微型鸡(WX) | 0.2145 | 0.2912 | 0.1177 | 0.1693 |
| 仙居鸡(XJ) | 0.1834 | 0.2755 | 0.1350 | 0.1735 |
| 萧山鸡(XS) | 0.2096 | 0.283 | 0.1470 | 0.1779 |
| 藏鸡(ZZ) | 0.1983 | 0.2961 | 0.1413 | 0.1659 |
Table 3
Correlation and fitting analysis of two groups of genetic diversity statistics"
| 统计量 | 皮尔逊相关系数 | 拟合曲线 | 拟合度R2 |
|---|---|---|---|
| 杂合度(Ho) | 0.794** | y=0.12×15.71x | 0.644*** |
| 近交系数(FROH) | 0.926** | y=0.17-4.69x+50.97x2-132.84x3 | 0.916*** |
| 多态信息含量(PIC) | 0.901** | y=0.43-1.79x+4.48x2 | 0.827*** |
| 平均遗传分化系数(Fst) | 0.984** | 0.972*** |
| [1] | Vekić M, Kalamujić Stroil B, Trivunović S, Pojskić N, Djukić Stojčić M. Genetic diversity of Banat Naked Neck, indigenous chicken breed from Serbia, inferred from mitochondrial DNA D-loop sequence and microsatellite markers. Anim Biotechnol, 2023, 34(7): 2197-2206. |
| [2] |
Andrews KR, Good JM, Miller MR, Luikart G, Hohenlohe PA. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet, 2016, 17(2): 81-92.
doi: 10.1038/nrg.2015.28 pmid: 26729255 |
| [3] | Zhang KF, Zhou YD, Song WH, Jiang LH, Yan XJ. Genome-wide RADseq reveals genetic differentiation of wild and cultured populations of large yellow croaker. Genes (Basel), 2023, 14(7): 1508. |
| [4] |
Clugston JAR, Kenicer GJ, Milne R, Overcast I, Wilson TC, Nagalingum NS. RADseq as a valuable tool for plants with large genomes-A case study in cycads. Mol Ecol Resour, 2019, 19(6): 1610-1622.
doi: 10.1111/1755-0998.13085 pmid: 31484214 |
| [5] | Zhu YF, Yin JM, Zhang JF, Li GH, Shen HY, Xue Q, Zhang HY, DOU XH, Su YJ, Han W. Genomic selection signal analysis of Langshan chicken based on RAD-seq sequencing. China Anim Husb Vet Med, 2020, 47(9): 2706-2714. |
|
朱云芬, 殷建玫, 张吉发, 李国辉, 沈海玉, 薛倩, 张会永, 窦新红, 苏一军, 韩威. 基于RAD-seq测序的狼山鸡基因组选择信号分析. 中国畜牧兽医, 2020, 47(9): 2706-2714.
doi: 10.16431/j.cnki.1671-7236.2020.09.002 |
|
| [6] | Yin JM, Zhu YF, Li GH, Zhang HY, Xue Q, Zhou CH, Jiang YX, Su YJ, Han W. Genetic evolution of Langya chicken based on simplified genome sequencing based on RAD-Seq. China Poult, 2023, 45(5): 19-23. |
| 殷建玫, 朱云芬, 李国辉, 张会永, 薛倩, 周成浩, 蒋一秀, 苏一军, 韩威. 基于RAD-Seq简化基因组测序分析琅琊鸡的遗传进化. 中国家禽, 2023, 45(5): 19-23. | |
| [7] | Ma LX, Cao GW, Zhu HF, Deng ZZ, Cai ZY, Zhou CH, Han W, Gu YL, Zhang J. Analysis of genetic variation based on RAD-seq breeding population of Jingyuan chickens. Acta Vet Zootech Sin, 2022, 53(7): 2104-2117. |
|
马丽霞, 曹国伟, 朱红芳, 邓占钊, 蔡正云, 周成浩, 韩威, 顾亚玲, 张娟. 基于RAD-seq静原鸡保种群体的遗传变异分析. 畜牧兽医学报, 2022, 53(7): 2104-2117.
doi: 10.11843/j.issn.0366-6964.2022.07.008 |
|
| [8] | Han W, Zhu YF, Yin JM, Li GH, Xue Q, Zhang HY, Shen HY, Su YJ, Dou XH, Wang KH, Zou JM. Genetic evolution of 19 local chicken breeds based on simplified genome sequencing by RAD-seq. Acta Vet Zootech Sin, 2020, 51(4): 670-678. |
|
韩威, 朱云芬, 殷建玫, 李国辉, 薛倩, 张会永, 沈海玉, 苏一军, 窦新红, 王克华, 邹剑敏. 基于RAD-seq简化基因组测序的19个地方鸡种遗传进化研究. 畜牧兽医学报, 2020, 51(4): 670-678.
doi: 10.11843/j.issn.0366-6964.2020.04.003 |
|
| [9] | Wang F, Guo YW, Liu ZY, Wang Q, Jiang Y, Zhao GP. New insights into the novel sequences of the chicken pan-genome by liquid chip. J Anim Sci, 2022, 100(12): skac336. |
| [10] |
Groenen MAM, Megens HJ, Zare Y, Warren WC, Hillier LW, Crooijmans RPMA, Vereijken A, Okimoto R, Muir WM, Cheng HH. The development and characterization of a 60K SNP chip for chicken. BMC Genomics, 2011, 12(1): 274.
doi: 10.1186/1471-2164-12-274 pmid: 21627800 |
| [11] |
Kranis A, Gheyas AA, Boschiero C, Turner F, Yu L, Smith S, Talbot R, Pirani A, Brew F, Kaiser P, Hocking PM, Fife M, Salmon N, Fulton J, Strom TM, Haberer G, Weigend S, Preisinger R, Gholami M, Qanbari S, Simianer H, Watson KA, Woolliams JA, Burt DW. Development of a high density 600K SNP genotyping array for chicken. BMC Genomics, 2013, 14: 59.
doi: 10.1186/1471-2164-14-59 pmid: 23356797 |
| [12] |
Liu RR, Xing SY, Wang J, Zheng MQ, Cui HX, Crooijmans RPMA, Li QH, Zhao GP, Wen J. A new chicken 55K SNP genotyping array. BMC Genomics, 2019, 20(1): 410.
doi: 10.1186/s12864-019-5736-8 pmid: 31117951 |
| [13] | Liu Z, Sun CJ, Yan YY, Li GQ, Li XC, Wu GQ, Yang N. Design and evaluation of a custom 50K Infinium SNP array for egg-type chickens. Poult Sci, 2021, 100(5): 101044. |
| [14] |
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet, 2007, 81(3): 559-575.
doi: 10.1086/519795 pmid: 17701901 |
| [15] | Serrote CML, Reiniger LRS, Silva KB, Rabaiolli SMDS, Stefanel CM. Determining the polymorphism information content of a molecular marker. Gene, 2020, 726: 144175. |
| [16] | Biscarini F, Mastrangelo S, Catillo G, Senczuk G, Ciampolini R. Insights into genetic diversity, runs of homozygosity and heterozygosity-rich regions in maremmana semi-feral cattle using pedigree and genomic data. Animals (Basel), 2020, 10(12): 2285. |
| [17] | Vahedi SM, Ardestani SS. FSTest: an efficient tool for cross-population fixation index estimation on variant call format files. J Genet, 2024, 103: 04. |
| [18] | Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One, 2010, 5(3): e9490. |
| [19] |
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol, 2021, 38(7): 3022-3027.
doi: 10.1093/molbev/msab120 pmid: 33892491 |
| [20] | Tian SS, Li W, Zhong ZQ, Wang FF, Xiao Q. Genome-wide re-sequencing data reveals the genetic diversity and population structure of Wenchang chicken in China. Anim Genet, 2023, 54(3): 328-337. |
| [21] | Tang XJ, Jia XX, Fan YF, Gao YS, Ge QL, Gu R, Lu JX, Liu YY, Wang J. Genetic diversity and phylogeny of Langya chickens based on mtDNA D-loop region. J Yangzhou Univ(Agric Life Sci Ed), 2017, 38(2): 47-51. |
| 唐修君, 贾晓旭, 樊艳凤, 高玉时, 葛庆联, 顾荣, 陆俊贤, 刘茵茵, 王珏. 基于mtDNA D-loop区全序列的琅琊鸡遗传多样性和系统进化研究. 扬州大学学报(农业与生命科学版), 2017, 38(2): 47-51. | |
| [22] | Tang XJ, Jia XX, Fan YF, Ge QL, Tang MJ, Chen DW, Zhang XY, Lu JX, Gao YS. Genetic polymorphism and origin of 4 chicken breeds based on Cytb gene complete sequence. Chin J Agric Biotechnol, 2020, 28(6): 83-91. |
| 唐修君, 贾晓旭, 樊艳凤, 葛庆联, 唐梦君, 陈大伟, 张小燕, 陆俊贤, 高玉时. 基于Cytb基因全序列4个鸡品种遗传多态性和起源. 农业生物技术学报, 2020, 28(6): 83-91. | |
| [23] |
Makanjuola BO, Maltecca C, Miglior F, Marras G, Abdalla EA, Schenkel FS, Baes CF. Identification of unique ROH regions with unfavorable effects on production and fertility traits in Canadian Holsteins. Genet Sel Evol, 2021, 53(1): 68.
doi: 10.1186/s12711-021-00660-z pmid: 34461820 |
| [24] |
Gorssen W, Meyermans R, Janssens S, Buys N. A publicly available repository of ROH islands reveals signatures of selection in different livestock and pet species. Genet Sel Evol, 2021, 53(1): 2.
doi: 10.1186/s12711-020-00599-7 pmid: 33397285 |
| [25] |
Grilz-Seger G, Druml T, Neuditschko M, Mesarič M, Cotman M, Brem G. Analysis of ROH patterns in the Noriker horse breed reveals signatures of selection for coat color and body size. Anim Genet, 2019, 50(4): 334-346.
doi: 10.1111/age.12797 pmid: 31199540 |
| [26] | Solmundson K, Bowman J, Manseau M, Taylor RS, Keobouasone S, Wilson PJ. Genomic population structure and inbreeding history of Lake Superior caribou. Ecol Evol, 2023, 13(7): e10278. |
| [27] | Fatma R, Chauhan W, Afzal M. The coefficients of inbreeding revealed by ROH study among inbred individuals belonging to each type of the first cousin marriage: a preliminary report from North India. Genes Genomics, 2023, 45(6): 813-825. |
| [28] | Di Gregorio P, Perna A, Di Trana A, Rando A. Identification of ROH islands conserved through generations in pigs belonging to the Nero Lucano breed. Genes (Basel), 2023, 14(7): 1503. |
| [29] | Chen KW, Li HF, Wang JY, Tang QP, Shen JC, Zhang SJ. Genetic variation of 27 local chicken breeds (lines) in East China. Acta Vet Zootech Sin, 2006, 37(1): 7-11. |
| 陈宽维, 李慧芳, 王金玉, 汤青萍, 沈见成, 章双杰. 华东27个地方鸡品种(品系)的遗传变异. 畜牧兽医学报, 2006, 37(1): 7-11. | |
| [30] |
Li HF, Han W, Zhu YF, Shu JT, Zhang XY, Chen KW. Analysis of genetic structure and relationship among nine indigenous Chinese chicken populations by the structure program. J Genet, 2009, 88(2): 197-203.
doi: 10.1007/s12041-009-0028-8 pmid: 19700858 |
| [31] | Zhuang Z, Zhao L, Zong WC, Guo QX, Li XF, Bi YL, Wang ZX, Jiang Y, Chen GH, Li BC, Chang GB, Bai H. Genetic diversity and breed identification of Chinese and Vietnamese local chicken breeds based on microsatellite analysis. J Anim Sci, 2023, 101: skad182. |
| [32] | Wang DQ, Chen GH, Wu XS, Zhang XY, Wang KH. Performance measurement and cluster analysis of main local chicken breeds in China. J Zhejiang Agric Sci, 2011, (4): 930-932. |
| 王德前, 陈国宏, 吴信生, 张学余, 王克华. 我国主要地方鸡品种性能测定与聚类分析. 浙江农业科学, 2011, (4): 930-932. |
| [1] | Jun Ma, Anping Fan, Wusheng Wang, Jinchuan Zhang, Xiaojun Jiang, Ruijun Ma, Sheqiang Jia, Fei Liu, Chuchao Lei, Yongzhen Huang. Analysis of genetic diversity and genetic structure of Qinchuan cattle conservation population using whole-genome resequencing [J]. Hereditas(Beijing), 2023, 45(7): 602-616. |
| [2] | Jie Kou, Yan Li, Peng Wang, Hong Liu, Jiawen Liu, Juan Wang, Ye Wang, Liang Zhang, Fujun Shen. Optimization of microsatellite genotyping system used for genetic diversity evaluation of Ailuropoda melanoleuca [J]. Hereditas(Beijing), 2022, 44(3): 253-266. |
| [3] | Haoyu Wang, Yuhan Hu, Yueyan Cao, Qiang Zhu, Yuguo Huang, Xi Li, Ji Zhang. AI-SNPs screening based on the whole genome data and research on genetic structure differences of subcontinent populations [J]. Hereditas(Beijing), 2021, 43(10): 938-948. |
| [4] | Zhiwei Xu, Yunlin Wei, Xiuling Ji. Progress on phage genomics of Pseudomonas spp. [J]. Hereditas(Beijing), 2020, 42(8): 752-759. |
| [5] | Zhi Li,Jun He,Jun Jiang,Richard G. Tait Jr.,Stewart Bauck,Wei Guo,Xiao-Lin Wu. Impacts of SNP genotyping call rate and SNP genotyping error rate on imputation accuracy inHolsteincattle [J]. Hereditas(Beijing), 2019, 41(7): 644-652. |
| [6] | Jianmin Zheng,Jiangtao Luo,Hongshen Wan,Shizhao Li,Manyu Yang,Jun Li,Ennian Yang,Yun Jiang,Yubin Liu,Xiangquan Wang,Zongjun Pu. Pedigree and development of wheat varieties in Sichuan Province [J]. Hereditas(Beijing), 2019, 41(7): 599-610. |
| [7] | Jun He,Changsong Qian,Richard G. Tait Jr.,Stewart Bauck,Xiaolin Wu. Estimating genomic breed composition of individual animals using selected SNPs [J]. Hereditas(Beijing), 2018, 40(4): 305-314. |
| [8] | Zhao Yongxin, Li Menghua. Research advances on the origin, evolution and genetic diversity of Chinese native sheep breeds [J]. Hereditas(Beijing), 2017, 39(11): 958-973. |
| [9] | Xian Gong, Chao Zhang, Aisa Yiliyasi, Ying Shi, Xuewei Yang, Aosiman Nuersimanguli, Yaqun Guan, Shuhua Xu. A comparative analysis of genetic diversity of candidate genes associated with type 2 diabetes in worldwide populations [J]. Hereditas(Beijing), 2016, 38(6): 543-559. |
| [10] | HUANG Yi-Min XIA Meng-Ying HUANG Shi. Evolutionary process unveiled by the maximum genetic diversity hypothesis [J]. HEREDITAS, 2013, 35(5): 599-606. |
| [11] | WEN Ying LU Xiao-Ping REN Rui MI Fu-Gui HAN Ping-An XUE Chun-Lei. Development of EST-SSR marker and genetic diversity analysis in Sorghum bicolor×Sorghum sudanenes [J]. HEREDITAS, 2013, 35(2): 225-232. |
| [12] | LI Duo CHAI Zhi-Xin JI Qiu-Mei ZHANG Cheng-Fu XIN Jin-Wei ZHONG Jin-Cheng. Genetic diversity of DNA microsatellite for Tibetan Yak [J]. HEREDITAS, 2013, 35(2): 175-184. |
| [13] | MA Zhi-Jie ZHONG Jin-Cheng HAN Jian-Lin XU Jing-Tao LIU Zhong-Na BAI Wen-Lin. Research progress on molecular genetic diversity of the yak (Bos grunniens) [J]. HEREDITAS, 2013, 35(2): 151-160. |
| [14] | FU Jian-Jun LI Jia-Le SHEN Yu-Bang WANG Rong-Quan XUAN Yun-Feng XU Xiao-Yan CHEN Yong. Genetic variation analysis of wild populations of grass carp (Ctenopharyngodon idella) using microsatellite markers [J]. HEREDITAS, 2013, 35(2): 192-201. |
| [15] | WANG Xiao-Qing WANG Chuan-Chao DENG Qiong-Ying LI Hui. Genetic analysis of Y chromosome and mitochondrial DNA poly-morphism of Mulam ethnic group in Guangxi, China [J]. HEREDITAS, 2013, 35(2): 168-174. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||