Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (8): 627-639.doi: 10.16288/j.yczz.24-137
• Review • Previous Articles Next Articles
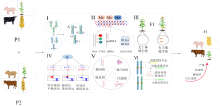
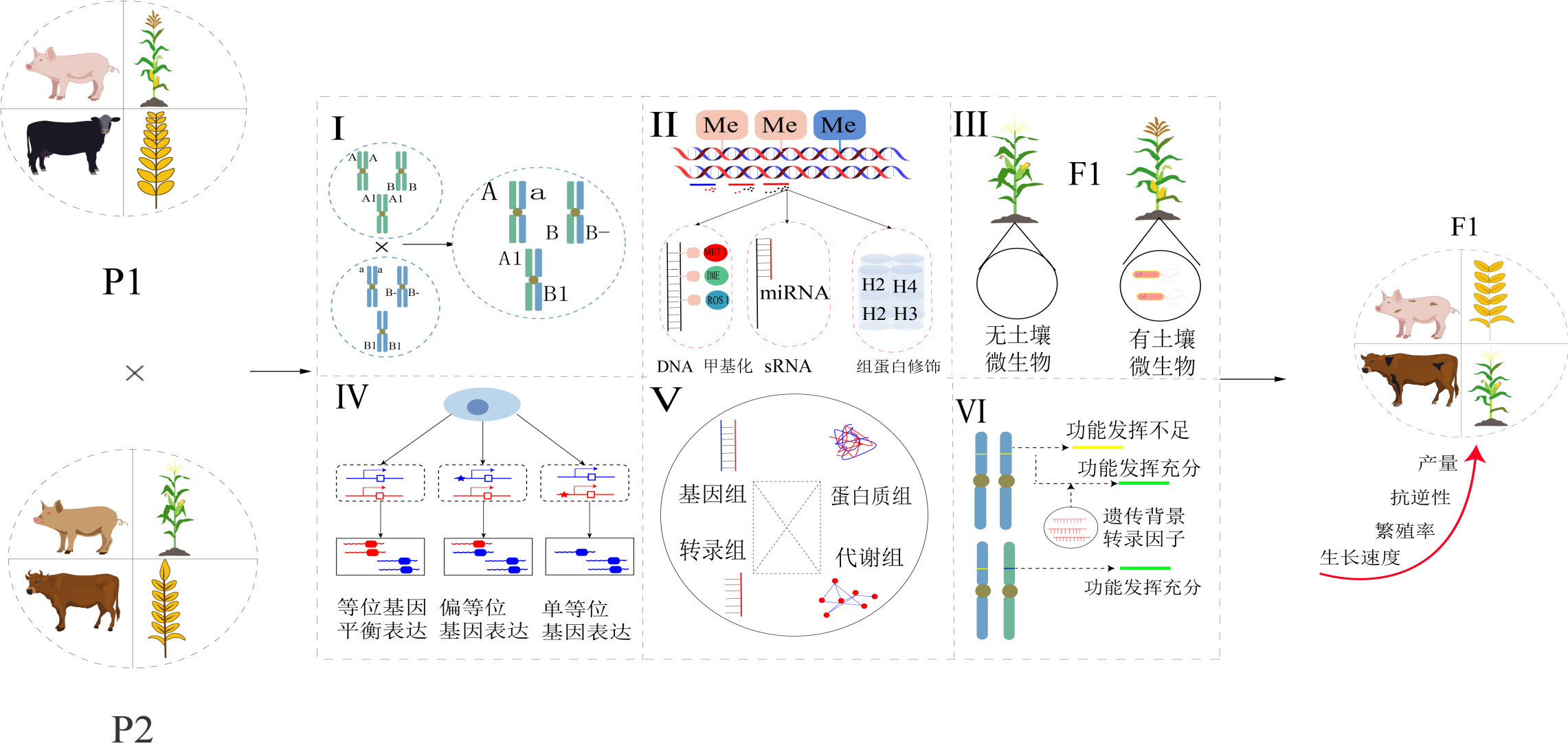
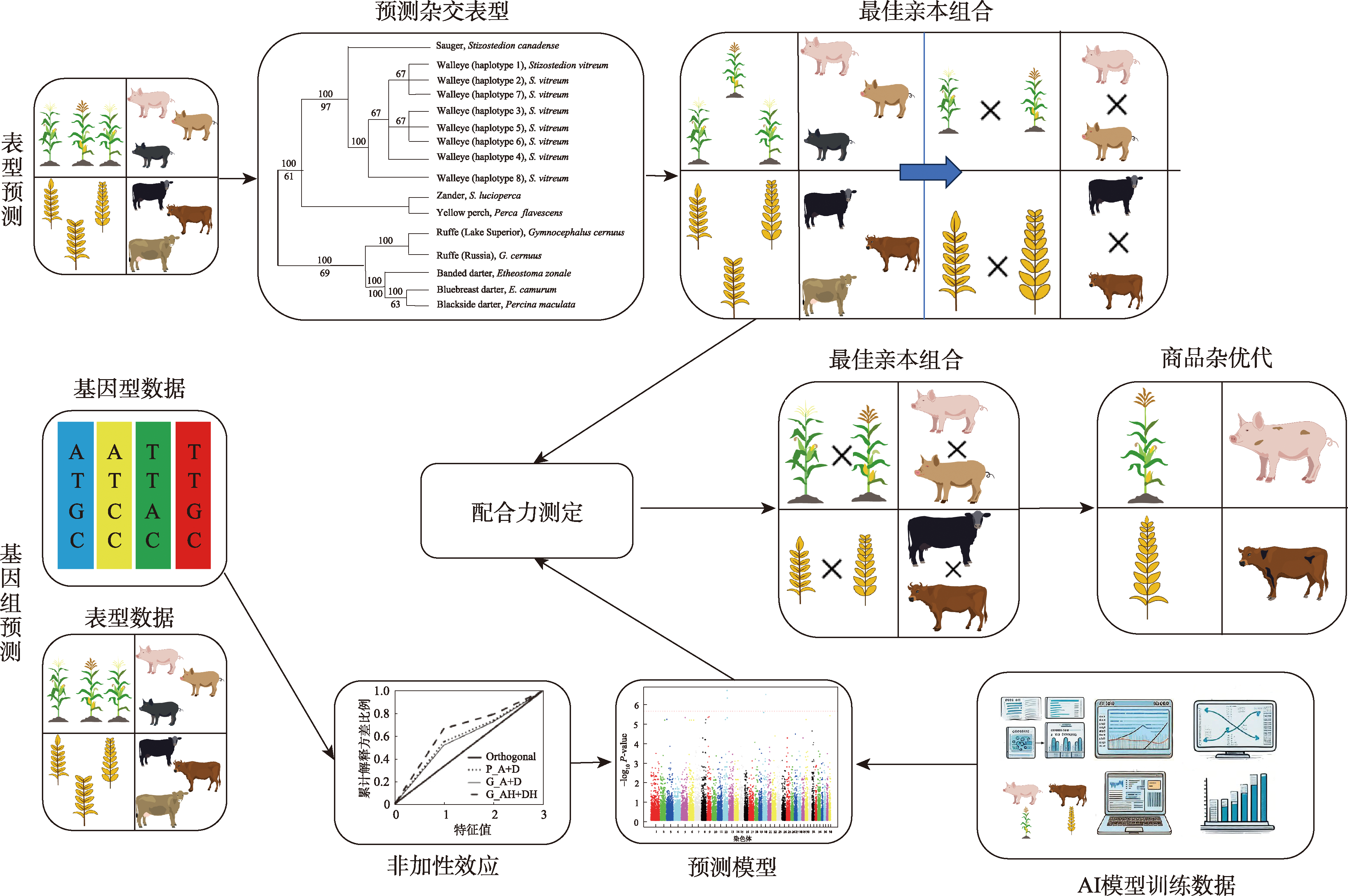
Heterosis formation mechanism, prediction methods, and their application and prospect in pig production
Jiahua Zhu( ), Junnan Shen, Xudong Yi, Ru Li, He Yu, Rongrong Ding, Weijun Pang(
), Junnan Shen, Xudong Yi, Ru Li, He Yu, Rongrong Ding, Weijun Pang( )
)
- Shaanxi Provincial Key Laboratory of Animal Genetics, Breeding and Reproduction, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China
-
Received:2024-05-11Revised:2024-07-03Online:2024-08-20Published:2024-07-12 -
Contact:Weijun Pang E-mail:zhujiahua@nwafu.edu.cn;pwj1226@nwafu.edu.cn -
Supported by:National Key Research and Development Program of China(2021YFF1000602);China Agriculture Research System of MOF and MARA(CARS-35);Key Research and Development Program of Shaanxi Province(2022ZDLNY01-04)
Cite this article
Jiahua Zhu, Junnan Shen, Xudong Yi, Ru Li, He Yu, Rongrong Ding, Weijun Pang. Heterosis formation mechanism, prediction methods, and their application and prospect in pig production[J]. Hereditas(Beijing), 2024, 46(8): 627-639.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Research progress on pig hybrid production"
| 杂交组合 | 主要研究性状 | 研究结果 | 参考文献 |
|---|---|---|---|
| 巴克夏猪、藏猪 | 胴体性状:皮重、瘦肉率、熟肉率等 肉质性状:氨基酸含量、脂肪酸含量等 | 巴藏猪瘦肉率比藏猪高21.2%,熟肉率高14.7%,必需氨基酸含量巴藏杂交猪比巴克夏猪高7%,非必需氨基酸高7% | [ |
| 高黎贡山猪、撒坝猪 | 生长性状:平均日增重、耗料增重比 胴体性状:瘦肉率、胴体重、平均背膘厚 | 高撒杂交猪的平均日增重比高黎贡山猪高5.9%,比撒坝猪低6.1%,胴体重比高黎贡山猪高39.2%,比撒坝猪低16% | [ |
| 巴克夏猪、青峪猪 | 繁殖性状:总出生数、活产数、窝出生体重等 肉质性状:屠宰率、滴水损失、肌肉pH值等 | 巴青猪比青峪猪重量提升18.54%,眼肌面积提升26.7%,45分钟失水率提升11.33% | [ |
| 塞尔塔猪、杜洛克猪、长白猪 | 生长性状:生长速度等 胴体性状:胴体长度、活重等 肉质性状:Fe-血红素等 | 杜塞猪与长塞猪比塞尔塔猪生长速度更快,杜塞猪比塞尔塔猪胴体长度长4.5%,两种杂交猪Fe-血红素比塞尔塔猪低33.51% | [ |
| 杜洛克、巴克夏、延安猪 | 胴体性状:胴体重、背膘厚 肉质性状:肌肉pH值、滴水损失、蒸煮损失 | 杂交猪胴体比延安猪重37.3%,杂交猪的滴水损失和蒸煮损失更低 | [ |
| 杜洛克、巴克夏、嘉兴黑猪 | 胴体性状:活重、眼肌面积等 肉质性状:肌苷酸、肌内脂肪、游离氨基酸含量等 | 胴体性状(活重、眼肌面积等)杜杜巴嘉杂交猪优于嘉兴黑猪,肉质性状嘉兴黑猪优于杂交猪。FLNC、DUSP4等介导脂肪生成肌肉增殖和分化 | [ |
| 巴克夏猪、广东小耳花猪 | 繁殖性状:总产仔数、产活仔数、初生窝重等 生长性状:平均日增重、耗料增重比等 | 巴花猪的初生窝重比广东小耳花猪高16.4%,巴花猪的平均日增重比广东小耳花猪高131.6% | [ |
| 杜洛克猪、长白猪、约克夏猪 | 肉质性状:pH值、肉色、大理石花纹等 | 杜长约杂交猪pH45值较亲本猪下降2%,肉色下降2% | [ |
| 嘉兴黑猪、巴克夏猪、杜洛克猪、长白猪 | 胴体性状:胴体重、活重、皮厚、背膘厚等 肉质性状:pH值、肉色、大理石花纹等 | 胴体重巴嘉、杜巴嘉、杜长嘉较嘉兴黑猪分别提升27.8%、62.7%和60%,大理石花纹评分杜×巴×嘉较嘉兴黑猪提升7% | [ |
| 本地猪(种丫杈猪、青峪猪、凉山猪、内江猪、成华猪、雅南猪)、长白猪、大白猪、川乡黑猪、杜洛克猪 | 胴体性状:瘦肉率、眼肌面积等 生长性状:平均日增重、平均耗料比等 | 90 kg时杜本猪、川本猪胴体瘦肉率比本地猪提高10%,川巴本三元杂交猪平均日增重比杜本、川本二元杂交猪提高约130g,平均耗料增重比下降约22% | [ |
| [1] | Han TW, Wang F, Song QX, Ye WX, Liu TS, Wang LM, Chen ZJ. An epigenetic basis of inbreeding depression in maize. Sci Adv, 2021, 7(35): eabg5442. |
| [2] |
Gu ZL, Gong JY, Zhu Z, Li Z, Feng Q, Wang CS, Zhao Y, Zhan QL, Zhou CC, Wang AH, Huang T, Zhang L, Tian QL, Fan DL, Lu YQ, Zhao Q, Huang XH, Yang SH, Han B. Structure and function of rice hybrid genomes reveal genetic basis and optimal performance of heterosis. Nat Genet, 2023, 55(10): 1745-1756.
doi: 10.1038/s41588-023-01495-8 pmid: 37679493 |
| [3] |
Xiao YJ, Jiang SQ, Cheng Q, Wang XQ, Yan J, Zhang RY, Qiao F, Ma C, Luo JY, Li WQ, Liu HJ, Yang WY, Song WH, Meng YJ, Warburton ML, Zhao JR, Wang XF, Yan JB. The genetic mechanism of heterosis utilization in maize improvement. Genome Biol, 2021, 22(1): 148.
doi: 10.1186/s13059-021-02370-7 pmid: 33971930 |
| [4] | Terada K, Ohtani T, Ogawa S, Hirooka H. Genetic parameters for carcass and meat quality traits in Jinhua, Duroc, and their crossbred pigs. J Anim Breed Genet, 2024, 141(1): 33-41. |
| [5] |
Bunning H, Wall E, Chagunda MGG, Banos G, Simm G. Heterosis in cattle crossbreeding schemes in tropical regions: meta-analysis of effects of breed combination, trait type, and climate on level of heterosis. J Anim Sci, 2019, 97(1): 29-34.
doi: 10.1093/jas/sky406 pmid: 30346552 |
| [6] | Hay EH, Roberts A. Genomic analysis of heterosis in an angus × hereford cattle population. Animals (Basel), 2023, 13(2): 191. |
| [7] |
Bruce AB. The Mendelian theory of heredity and the augmentation of vigor. Science, 1910, 32(827): 627-628.
doi: 10.1126/science.32.827.627-a pmid: 17816706 |
| [8] |
Jones DF. Dominance of linked factors as a means of accounting for heterosis. Proc Natl Acad Sci USA, 1917, 3(4): 310-312.
doi: 10.1073/pnas.3.4.310 pmid: 16586724 |
| [9] |
Hua JP, Xing YZ, Wu WR, Xu CG, Sun XL, Yu SB, Zhang QF. Single-locus heterotic effects and dominance by dominance interactions can adequately explain the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA, 2003, 100(5): 2574-2579.
pmid: 12604771 |
| [10] |
Hashimoto S, Wake T, Nakamura H, Minamiyama M, Araki-Nakamura S, Ohmae-Shinohara K, Koketsu E, Okamura S, Miura K, Kawaguchi H, Kasuga S, Sazuka T. The dominance model for heterosis explains culm length genetics in a hybrid sorghum variety. Sci Rep, 2021, 11(1): 4532.
doi: 10.1038/s41598-021-84020-3 pmid: 33633216 |
| [11] | Tang JH, Yan JB, Ma XQ, Teng WT, Wu WR, Dai JR, Dhillon BS, Melchinger AE, Li JS. Dissection of the genetic basis of heterosis in an elite maize hybrid by QTL mapping in an immortalized F2population. Theor Appl Genet, 2010, 120(2): 333-340. |
| [12] |
Khayatzadeh N, Mészáros G, Utsunomiya YT, Schmitz-Hsu F, Seefried F, Schnyder U, Ferenčaković M, Garcia JF, Curik I, Sölkner J. Genome-wide mapping of the dominance effects based on breed ancestry for semen traits in admixed Swiss Fleckvieh bulls. J Dairy Sci, 2019, 102(12): 11217-11224.
doi: S0022-0302(19)30834-3 pmid: 31548062 |
| [13] |
Cui LL, Yang B, Xiao SJ, Gao J, Baud A, Graham D, Mcbride M, Dominiczak A, Schafer S, Aumatell RL, Mont C, Teruel AF, Hübner N, Flint J, Mott R, Huang LS. Dominance is common in mammals and is associated with trans-acting gene expression and alternative splicing. Genome Biol, 2023, 24(1): 215.
doi: 10.1186/s13059-023-03060-2 pmid: 37773188 |
| [14] |
Birchler JA, Auger DL, Riddle NC. In search of the molecular basis of heterosis. Plant Cell, 2003, 15(10): 2236-2239.
doi: 10.1105/tpc.151030 pmid: 14523245 |
| [15] | Shull GH. The composition of a field of maize. J Hered, 1908, 4(1): 296-301. |
| [16] | Kaeppler SM. Heterosis: many genes, many mechanisms—end the search for an undiscovered unifying theory. Inter Schol Res Not, 2012: 682824. |
| [17] |
Liu HJ, Wang Q, Chen MJ, Ding YH, Yang XR, Liu J, Li XH, Zhou CC, Tian QL, Lu YQ, Fan DL, Shi JP, Zhang L, Kang CB, Sun MF, Li FY, Wu YJ, Zhang YZ, Liu BS, Zhao XY, Feng Q, Yang JL, Han B, Lai JS, Zhang XS, Huang XH. Genome-wide identification and analysis of heterotic loci in three maize hybrids. Plant Biotechnol J, 2020, 18(1): 185-194.
doi: 10.1111/pbi.13186 pmid: 31199059 |
| [18] |
Lin ZC, Qin P, Zhang XW, Fu CJ, Deng HC, Fu XX, Huang Z, Jiang SQ, Li C, Tang XY, Wang XF, He GM, Yang YZ, He H, Deng XW. Divergent selection and genetic introgression shape the genome landscape of heterosis in hybrid rice. Proc Natl Acad Sci USA, 2020, 117(9): 4623-4631.
doi: 10.1073/pnas.1919086117 pmid: 32071222 |
| [19] | Shalby N, Mohamed IAA, Xiong J, Hu KN, Yang YBT, Nishawy E, Yi B, Wen J, Ma CZ, Shen JX, Fu TD, Tu JX. Overdominance at the gene expression level plays a critical role in the hybrid root growth of Brassica napus. Int J Mol Sci, 2021, 22(17): 9246. |
| [20] | Torgeman S, Zamir D. Epistatic QTLs for yield heterosis in tomato. Proc Natl Acad Sci USA, 2023, 120(14): e2205787119. |
| [21] | Tian SH, Xu XL, Zhu XF, Wang F, Song XL, Zhang TZ. Overdominance is the major genetic basis of lint yield heterosis in interspecific hybrids between G. hirsutum and G. barbadense. Heredity (Edinb), 2019, 123(3): 384-394. |
| [22] | Xiao W, Chen BL, Wang J, Zou ZY, Wang CH, Li DY, Zhu JL, Yu J, Yang H. Integration of mRNA and miRNA profiling reveals heterosis in Oreochromis niloticus × O. aureus hybrid tilapia. Animals (Basel), 2022, 12(5): 640. |
| [23] | Wallace JG, Larsson SJ, Buckler ES. Entering the second century of maize quantitative genetics. Heredity (Edinb), 2014, 112(1): 30-38. |
| [24] |
Bateson W. Facts limiting the theory of heredity. Science, 1907, 26(672): 649-660.
pmid: 17796786 |
| [25] | Li H, Jiang SQ, Li C, Liu L, Lin ZC, He H, Deng XW, Zhang ZD, Wang XF. The hybrid protein interactome contributes to rice heterosis as epistatic effects. Plant J, 2020, 102(1): 116-128. |
| [26] | Boeven PHG, Zhao YS, Thorwarth P, Liu F, Maurer HP, Gils M, Schachschneider R, Schacht J, Ebmeyer E, Kazman E, Mirdita V, Dörnte J, Kontowski S, Horbach R, Cöster H, Holzapfel J, Jacobi A, Ramgraber L, Reinbrecht C, Starck N, Varenne P, Starke A, Schürmann F, Ganal M, Polley A, Hartung J, Beier S, Scholz U, Longin CFH, Reif JC, Jiang Y, Würschum T. Negative dominance and dominance-by-dominance epistatic effects reduce grain-yield heterosis in wide crosses in wheat. Sci Adv, 2020, 6(24): eaay4897. |
| [27] | Sang ZQ, Wang H, Yang YX, Zhang ZQ, Liu XG, Li ZW, Xu YB. Epistasis activation contributes substantially to heterosis in temperate by tropical maize hybrids. Front Plant Sci, 2022, 11(13): 921608. |
| [28] |
Wang C, Liu Q, Shen Y, Hua YF, Wang JJ, Lin JR, Wu MG, Sun TT, Cheng ZK, Mercier R, Wang KJ. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat Biotechnol, 2019, 37(3): 283-286.
doi: 10.1038/s41587-018-0003-0 pmid: 30610223 |
| [29] |
Nakamura S, Hosaka K. DNA methylation in diploid inbred lines of potatoes and its possible role in the regulation of heterosis. Theor Appl Genet, 2010, 120(2): 205-214.
doi: 10.1007/s00122-009-1058-6 pmid: 19455300 |
| [30] | He XJ, Ma ZY, Liu ZW. Non-coding RNA transcription and RNA-directed DNA methylation in Arabidopsis. Mol Plant, 2014, 7(9): 1406-1414. |
| [31] | Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, McCombie WR, Lavine K, Mittal V, May B, Kasschau KD, Carrington JC, Doerge RW, Colot V, Martienssen R. Role of transposable elements in heterochromatin and epigenetic control. Nature, 2004, 430(6998): 471-476. |
| [32] |
Yang L, Liu PT, Wang XC, Jia AL, Ren DQ, Tang YR, Tang YQ, Deng XW, He GM. A central circadian oscillator confers defense heterosis in hybrids without growth vigor costs. Nat Commun, 2021, 12(1): 2317.
doi: 10.1038/s41467-021-22268-z pmid: 33875651 |
| [33] |
Zhou SR, Xing MQ, Zhao ZL, Gu YC, Xiao YP, Liu QQ, Xue HW. DNA methylation modification in heterosis initiation through analyzing rice hybrid contemporary seeds. Crop J, 2021, 9(5): 1179-1190.
doi: 10.1016/j.cj.2020.12.003 |
| [34] | Chen LY, Zhu YY, Ren XB, Yao D, Song Y, Fan SJ, Li XY, Zhang Z, Yang SN, Zhang J, Zhang J. Heterosis and differential DNA methylation in soybean hybrids and their parental lines. Plants (Basel), 2022, 11(9): 1136. |
| [35] | Yang H, Li Q. The DNA methylation level is associated with the superior growth of the hybrid crosses in the Pacific oyster Crassostrea gigas. Aquaculture, 2022, 547: 737421. |
| [36] | Chen BW, Yue YJ, Li JY, Yuan C, Guo TT, Zhang D, Liu JB, Yang BH, Lu ZK. Global DNA methylation, miRNA, and mRNA profiles in sheep skeletal muscle promoted by hybridization. J Agric Food Chem, 2023, 71(41): 15398-15406. |
| [37] |
Greaves IK, Groszmann M, Ying H, Taylor JM, Peacock WJ, Dennis ES. Trans chromosomal methylation in Arabidopsis hybrids. Proc Natl Acad Sci USA, 2012, 109(9): 3570-3575.
doi: 10.1073/pnas.1201043109 pmid: 22331882 |
| [38] | Shen HS, He H, Li JG, Chen W, Wang XC, Guo L, Peng ZY, He GM, Zhong SW, Qi YJ, Terzaghi W, Deng XW. Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell, 2012, 24(3): 875-892. |
| [39] | Kawanabe T, Ishikura S, Miyaji N, Sasaki T, Wu LM, Itabashi E, Takada S, Shimizu M, Takasaki-Yasuda T, Osabe K, Peacock WJ, Dennis ES, Fujimoto R. Role of DNA methylation in hybrid vigor in Arabidopsis thaliana. Proc Natl Acad Sci USA, 2016, 113(43): E6704-E6711. |
| [40] |
Zhang QZ, Li YQ, Xu T, Srivastava AK, Wang D, Zeng L, Yang L, He L, Zhang H, Zheng ZM, Yang DL, Zhao C, Dong J, Gong ZZ, Liu RY, Zhu JK. The chromatin remodeler DDM1 promotes hybrid vigor by regulating salicylic acid metabolism. Cell Discov, 2016, 2: 16027.
doi: 10.1038/celldisc.2016.27 pmid: 27551435 |
| [41] | Luo JH, Wang M, Jia GF, He Y. Transcriptome-wide analysis of epitranscriptome and translational efficiency associated with heterosis in maize. J Exp Bot, 2021, 72(8): 2933-2946. |
| [42] | Dahal D, Mooney BP, Newton KJ. Specific changes in total and mitochondrial proteomes are associated with higher levels of heterosis in maize hybrids. Plant J, 2012, 72(1): 70-83. |
| [43] |
Zhang L, Peng YG, Wei XL, Dai Y, Yuan DW, Lu YF, Pan YY, Zhu Z. Small RNAs as important regulators for the hybrid vigour of super-hybrid rice. J Exp Bot, 2014, 65(20): 5989-6002.
doi: 10.1093/jxb/eru337 pmid: 25129133 |
| [44] |
Hill PWS, Amouroux R, Hajkova P. DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story. Genomics, 2014, 104(5): 324-333.
doi: 10.1016/j.ygeno.2014.08.012 pmid: 25173569 |
| [45] |
Gu HC, Qi X, Jia YX, Zhang ZB, Nie CS, Li XH, Li JY, Jiang ZH, Wang Q, Qu LJ. Inheritance patterns of the transcriptome in hybrid chickens and their parents revealed by expression analysis. Sci Rep, 2019, 9(1): 5750.
doi: 10.1038/s41598-019-42019-x pmid: 30962479 |
| [46] | Li DJ, Zeng RZ, Li Y, Zhao MM, Chao JQ, Li Y, Wang K, Zhu LH, Tian WM, Liang CZ. Gene expression analysis and SNP/InDel discovery to investigate yield heterosis of two rubber tree F1 hybrids. Sci Rep, 2016, 6(1): 24984. |
| [47] | Fu J, Zhang YL, Yan TZ, Li YF, Jiang N, Zhou YB, Zhou QF, Qin P, Fu CJ, Lin HY, Zhong J, Han X, Lin ZC, Wang F, He H, Wang K, Yang YZ. Transcriptome profiling of two super hybrid rice provides insights into the genetic basis of heterosis. Bmc Plant Biol, 2022, 22(1): 314. |
| [48] | Guo HB, Mendrikahy JN, Xie L, Deng JF, Lu ZJ, Wu JW, Li X, Shahid MQ, Liu XD. Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci Rep, 2017, 7(1): 40139. |
| [49] | Li PR, Su TB, Zhang DS, Wang WH, Xin XY, Yu YJ, Zhao XY, Yu SC, Zhang FL. Genome-wide analysis of changes in miRNA and target gene expression reveals key roles in heterosis for Chinese cabbage biomass. Hortic Res, 2021, 8(1): 39. |
| [50] |
Sun D, Wang D, Zhang Y, Yu Y, Xu G, Li J. Differential gene expression in liver of inbred chickens and their hybrid offspring. Anim genet, 2005, 36(3): 210-215.
pmid: 15932399 |
| [51] |
Xie JY, Wang WP, Yang T, Zhang Q, Zhang ZF, Zhu XY, Li N, Zhi LR, Ma XQ, Zhang SY, Liu Y, Wang XQ, Li FM, Zhao Y, Jia XW, Zhou JY, Jiang NJ, Li GL, Liu MS, Liu SJ, Li L, Zeng A, Du MK, Zhang ZY, Li JJ, Zhang ZD, Li ZC, Zhang HL. Large-scale genomic and transcriptomic profiles of rice hybrids reveal a core mechanism underlying heterosis. Genome Biol, 2022, 23(1): 264.
doi: 10.1186/s13059-022-02822-8 pmid: 36550554 |
| [52] | Wagner MR, Tang C, Salvato F, Clouse KM, Bartlett A, Vintila S, Phillips L, Sermons S, Hoffmann M, Balint-Kurti PJ, Kleiner M. Microbe-dependent heterosis in maize. Proc Natl Acad Sci USA, 2021, 118(30): e2021965118. |
| [53] | Wang XM, Zhou TH, Li GW, Yao W, Hu W, Wei X, Che J, Yang HC, Shao L, Hua JP, Li XH, Xiao JH, Xing YZ, Ouyang YD, Zhang QF. A Ghd7-centered regulatory network provides a mechanistic approximation to optimal heterosis in an elite rice hybrid. Plant J, 2022, 112(1): 68-83. |
| [54] |
Herbst RH, Bar-Zvi D, Reikhav S, Soifer I, Breker M, Jona G, Shimoni E, Schuldiner M, Levy AA, Barkai N. Heterosis as a consequence of regulatory incompatibility. BMC biol, 2017, 15(1): 38.
doi: 10.1186/s12915-017-0373-7 pmid: 28494792 |
| [55] | Wei XZ, Zhang JZ. The optimal mating distance resulting from heterosis and genetic incompatibility. Sci Adv, 2018, 4(11): eaau5518. |
| [56] | Pandey SK, Dasgupta T, Rathore A, Vemula A. Relationship of parental genetic distance with heterosis and specific combining ability in sesame (Sesamum indicum L.) based on phenotypic and molecular marker analysis. Biochem Genet, 2018, 56(3): 188-209. |
| [57] | Kawamura K, Kawanabe T, Shimizu M, Nagano AJ, Saeki N, Okazaki K, Kaji M, Dennis ES, Osabe K, Fujimoto R. Genetic distance of inbred lines of Chinese cabbage and its relationship to heterosis. Plant Gene, 2016, 5: 1-7. |
| [58] | Zhao ML, Han B, Willms WD. Detection of genetic diversity in rough fescue (Festuca campestris Rydb) populations of southern Alberta and British Columbia, Canada, using RAPD markers. Can. J. Plant Sci, 2008, 88(2): 307-312. |
| [59] | Yang YY, He RQ, Zheng J, Hu ZH, Wu J, Leng PS. Development of EST-SSR markers and association mapping with floral traits in Syringa oblata. Bmc Plant Biol, 2020, 20(1): 436. |
| [60] |
Li CT, Zhang SH, Li L, Chen JZ, Liu Y, Zhao SM. Selection of 29 highly informative InDel markers for human identification and paternity analysis in Chinese Han population by the SNPlex genotyping system. Mol Biol Rep, 2012, 39(3): 3143-3152.
doi: 10.1007/s11033-011-1080-z pmid: 21681421 |
| [61] | Lin TZ, Zhou C, Chen GM, Yu J, Wu W, Ge YW, Liu XL, Li J, Jiang XZ, Tang WJ, Tian YL, Zhao ZG, Zhu CS, Wang CM, Wan JM. Heterosis-associated genes confer high yield in super hybrid rice. Theor Appl Genet, 2020, 133(12): 3287-3297. |
| [62] | Wang BB, Hou M, Shi JP, Ku LX, Song W, Li CH, Ning Q, Li X, Li CY, Zhao BB, Zhang RY, Xu H, Bai ZJ, Xia ZC, Wang H, Kong DX, Wei HB, Jing YF, Dai ZY, Wang HH, Zhu XY, Sun X, Wang SS, Yao W, Hou GG, Qi Z, Dai H, Li XM, Zheng HK, Zhang ZX, Li Y, Wang TY, Jiang TJ, Wan ZM, Chen YH, Zhao JR, Lai JS, Wang HY. De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis. Nat Genet, 2023, 55(2): 312-323. |
| [63] | Luo X, Ma CZ, Yi B, Tu JX, Shen JX, Fu TD. Genetic distance revealed by genomic single nucleotide polymorphisms and their relationships with harvest index heterotic traits in rapeseed (Brassica napus L.). Euphytica, 2016, 209(1): 41-47. |
| [64] |
Knoch D, Werner CR, Meyer RC, Riewe D, Abbadi A, Lücke S, Snowdon RJ, Altmann T. Multi-omics-based prediction of hybrid performance in canola. Theor Appl Genet, 2021, 134(4): 1147-1165.
doi: 10.1007/s00122-020-03759-x pmid: 33523261 |
| [65] |
Seifert F, Thiemann A, Schrag TA, Rybka D, Melchinger AE, Frisch M, Scholten S. Small RNA-based prediction of hybrid performance in maize. BMC Genomics, 2018, 19(1): 371.
doi: 10.1186/s12864-018-4708-8 pmid: 29783940 |
| [66] |
Varona L, Legarra A, Toro MA, Vitezica ZG. Non-additive effects in genomic selection. Front Genet, 2018, 9: 78.
doi: 10.3389/fgene.2018.00078 pmid: 29559995 |
| [67] |
Moghaddar N, van der Werf JHJ. Genomic estimation of additive and dominance effects and impact of accounting for dominance on accuracy of genomic evaluation in sheep populations. J Anim Breed Genet, 2017, 134(6): 453-462.
doi: 10.1111/jbg.12287 pmid: 28833716 |
| [68] |
Heidaritabar M, Wolc A, Arango J, Zeng J, Settar P, Fulton JE, O'Sullivan NP, Bastiaansen JWM, Fernando RL, Garrick DJ, Dekkers JCM. Impact of fitting dominance and additive effects on accuracy of genomic prediction of breeding values in layers. J Anim Breed Genet, 2016, 133(5): 334-346.
doi: 10.1111/jbg.12225 pmid: 27357473 |
| [69] |
Esfandyari H, Bijma P, Henryon M, Christensen OF, Sørensen AC. Genomic prediction of crossbred performance based on purebred Landrace and Yorkshire data using a dominance model. Genet Sel Evol, 2016, 48(1): 40.
doi: 10.1186/s12711-016-0220-2 pmid: 27276993 |
| [70] |
Martini JWR, Wimmer V, Erbe M, Simianer H. Epistasis and covariance: how gene interaction translates into genomic relationship. Theor Appl Genet, 2016, 129(5): 963-976.
doi: 10.1007/s00122-016-2675-5 pmid: 26883048 |
| [71] | Clasen JB, Fikse WF, Su G, Karaman E. Multibreed genomic prediction using summary statistics and a breed-origin-of-alleles approach. Heredity (Edinb), 2023, 131(1): 33-42. |
| [72] |
Stock J, Bennewitz J, Hinrichs D, Wellmann R. A Review of genomic models for the analysis of livestock crossbred data. Front Genet, 2020, 11: 568.
doi: 10.3389/fgene.2020.00568 pmid: 32670349 |
| [73] | Hayes BJ, Chen CS, Powell O, Dinglasan E, Villiers K, Kemper KE, Hickey LT. Advancing artificial intelligence to help feed the world. Nat Biotechnol, 2023, 41(9): 1188-1189. |
| [74] | Luo J, Shen LY, Tan ZD, Cheng X, Yang DL, Fan Y, Yang Q, Ma JD, Tang QZ, Jiang AA, Jiang DM, Tang GQ, Jiang YZ, Li XW, Yang RL, Zhang SH, Zhu L. Comparison reproductive, growth performance, carcass and meat quality of Liangshan pig crossbred with Duroc and Berkshire genotypes and heterosis prediction. Livest Sci, 2018, 212: 61-68. |
| [75] | Chen C, Zhu J, Ren HB, Deng Y, Zhang X, Liu YY, Cui QM, Hu XG, Zuo JB, Chen B, Zhang X, Wu MS, Peng YL. Growth performance, carcass characteristics, meat quality and chemical composition of the Shaziling pig and its crossbreeds. Livest Sci, 2021, 244: 104342. |
| [76] | Lu FJ, Hao T, Liu QY, Gao Y, Ma ZC, Wang ZD, Zhang Q, Li N. Carcass performance and meat quality analysis of Berkshire pigs, Tibetan pigs and hybrid pigs. Chin J Anim Sci, 2024, 60(1): 240-243, 392. |
| 鹿富俊, 郝桐, 刘庆雨, 高一, 马峥财, 王正丹, 张庆, 李娜. 巴克夏猪、藏猪和巴藏杂交猪的胴体性能及肉品质分析. 中国畜牧杂志, 2024, 60(1): 240-243, 392. | |
| [77] | Zhou QB, He S, Zhao GS, He R, Wu LX, Yang ST, Liu JL, Yu SH, Zhao GY. Comparative analysis of production performance between Gao × Sa crossbred pigs and Gaoligongshan pigs and Saba pigs. Chin J Anim Sci, 2023, 59(8): 174-178. |
| 周戚斌, 和胜, 赵国松, 何荣, 吴玲想, 杨世婷, 刘珈纶, 余世海, 赵桂英. 高×撒杂交猪与高黎贡山猪和撒坝猪生产性能的比较分析. 中国畜牧杂志, 2023, 59(8): 174-178. | |
| [78] | Luo J, Yang YT, Liao K, Liu B, Chen Y, Shen LY, Chen L, Jiang AA, Liu YH, Li Q, Wang JY, Li XW, Zhang SH, Zhu L. Genetic parameter estimation for reproductive traits in QingYu pigs and comparison of carcass and meat quality traits to Berkshire × QingYu crossbred pigs. Asian-Australas J Anim Sci, 2020, 33(8): 1224-1232. |
| [79] |
Franco D, Vazquez JA, Lorenzo JM. Growth performance, carcass and meat quality of the Celta pig crossbred with Duroc and Landrance genotypes. Meat Sci, 2014, 96(1): 195-202.
doi: 10.1016/j.meatsci.2013.06.024 pmid: 23896154 |
| [80] |
Chen Y, Wei YY, Chen JN, Lv DJ, Li PN, Zhu L, Tang G, Li XW, Jiang YZ. Growth, carcass characteristics and meat quality of Chinese indigenous Yanan pig crossbred with Duroc and Berkshire genotypes. Anim Prod Sci, 2019, 59(6): 1147-1154.
doi: 10.1071/AN17450 |
| [81] | Chen QQ, Zhang W, Cai JF, Ni YF, Xiao LX, Zhang JZ. Transcriptome analysis in comparing carcass and meat quality traits of Jiaxing Black pig and Duroc × Duroc × Berkshire × Jiaxing Black pig crosses. Gene, 2022, 808: 145978. |
| [82] | Li C, Wei MF, Mou DL, Chen JW, Wang YJ, Liu XH, Yang LF, Zeng JH. Analysis of hybridization effect between Berkshire and Guangdong Xiaoerhua pig. Chin J Anim Sci, 2019, 55(4): 62-65. |
| 李闯, 韦明飞, 莫德林, 陈建伟, 王永江, 刘小红, 阳林芳, 曾检华. 巴克夏与广东小耳花猪杂交效果分析. 中国畜牧杂志, 2019, 55(4): 62-65. | |
| [83] |
Zhang J, Chai J, Luo ZG, He H, Chen L, Liu XQ, Zhou QF. Meat and nutritional quality comparison of purebred and crossbred pigs. Anim Sci J. 2018, 89(1): 202-210.
doi: 10.1111/asj.12878 pmid: 28856768 |
| [84] |
Zhang W, Song QQ, Wu F, Zhang JZ, Xu MS, Li HH, Han ZJ, Gao HX, Xu NY. Evaluation of the four breeds in synthetic line of Jiaxing Black pigs and Berkshire for meat quality traits, carcass characteristics, and flavor substances. Anim Sci J, 2019, 90(4): 574-582.
doi: 10.1111/asj.13169 pmid: 30714281 |
| [85] | Tao X, Liang Y, Ying SC, Zhong ZJ, Yang XM, Lei YF, Gong JJ, Chen XH, Shu G, Lyu XB, Gu YR. Study on crossbreeding experiment of Chuanxiang black pig. Chin J Anim Sci, 2021, 57(z1): 214-217. |
| 陶璇, 梁艳, 应三成, 钟志君, 杨雪梅, 雷云峰, 龚建军, 陈晓晖, 舒刚, 吕学斌, 顾以韧. 川乡黑猪杂交试验研究. 中国畜牧杂志, 2021, 57(z1): 214-217. | |
| [86] | Jiang FY, Yin XF, Li ZW, Guo RJ, Wang J, Fan J, Zhang YD, Kang MS, Fan XM. Predicting heterosis via genetic distance and the number of SNPs in selected segments of chromosomes in maize. Front Plant Sci, 2023, 14: 1111961. |
| [87] | Yue LX, Zhang SJ, Zhang LK, Liu YJ, Cheng F, Li GL, Zhang SF, Zhang H, Sun RF, Li F. Heterotic prediction of hybrid performance based on genome-wide SNP markers and the phenotype of parental inbred lines in heading Chinese cabbage (Brassica rapa L. ssp. pekinensis). Sci Hortic, 2022, 296: 110907. |
| [88] | Fu CY, Ma C, Zhu MS, Liu WG, Ma XZ, Li JH, Liao YL, Liu DL, Gu XF, Wang HY, Wang F. Transcriptomic and methylomic analyses provide insights into the molecular mechanism and prediction of heterosis in rice. Plant J, 2023, 115(1): 139-154. |
| [89] | Wang Q, Yan T, Long ZB, Huang LNY, Zhu Y, Xu Y, Chen XY, Pak H, Li JQ, Wu DZ, Xu Y, Hua SJ, Jiang LX. Prediction of heterosis in the recent rapeseed (Brassica napus) polyploid by pairing parental nucleotide sequences. PLoS Genet, 2021, 17(11): e1009879. |
| [90] | Xu Z, Zhang Z, Sun H, Zhang XZ, Xu NY, Chen JC, Hua JQ, Zhong TM, Wang QS, Pan YC. Heterosis prediction of hybridization between Jinhua pig and Duroc, Landrace, Large White pig. Chin J Anim Sci, 2019, 55(1): 64-67. |
| 徐忠, 张哲, 孙浩, 张向喆, 徐宁迎, 陈究成, 华坚青, 钟土木, 王起山, 潘玉春. 金华猪与杜洛克、长白猪、大白猪杂交的杂种优势预测. 中国畜牧杂志, 2019, 55(1): 64-67. | |
| [91] | Wang YJ, Thakali K, Morse P, Shelby S, Chen JL, Apple J, Huang Y. Comparison of growth performance and meat quality traits of commercial cross-bred pigs versus the large black pig breed. Animals (Basel), 2021, 11(1): 200. |
| [92] |
Westhues M, Schrag TA, Heuer C, Thaller G, Utz HF, Schipprack W, Thiemann A, Seifert F, Ehret A, Schlereth A, Stitt M, Nikoloski Z, Willmitzer L, Schön CC, Scholten S, Melchinger AE. Omics-based hybrid prediction in maize. Theor Appl Genet, 2017, 130(9): 1927-1939.
doi: 10.1007/s00122-017-2934-0 pmid: 28647896 |
| [93] | Colantonio V, Ferrão LFV, Tieman DM, Bliznyuk N, Sims C, Klee HJ, Munoz P, Resende MFR. Metabolomic selection for enhanced fruit flavor. Proc Natl Acad Sci USA, 2022, 119(7): e2115865119. |
| [1] | Daiyuan Liu, Zhaohui Zhang, Xianjiang Kang. Research progress on the effect of sperm chromatin integrity on function and its detection methods [J]. Hereditas(Beijing), 2024, 46(7): 511-529. |
| [2] | Yuan Shen, Jintao Li, Miao Yin, Qunying Lei. The roles of branched-chain amino acids metabolism in tumorigenesis and progression [J]. Hereditas(Beijing), 2024, 46(6): 438-451. |
| [3] | Zhaoran Sun, Xudong Wu. The roles and mechanisms of histone variant H2A.Z in transcriptional regulation [J]. Hereditas(Beijing), 2024, 46(4): 279-289. |
| [4] | Zhenlin Cao, Jinhong Li, Minhui Zhou, Manting Zhang, Ning Wang, Yifei Chen, Jiaxin Li, Qingsong Zhu, Wenjun Gong, Xuchen Yang, Xiaolong Fang, Jiaxian He, Meina Li. Functional study of the soybean stamen-preferentially expressed gene GmFLA22a in regulating male fertility [J]. Hereditas(Beijing), 2024, 46(4): 333-345. |
| [5] | Xiangdong Liu, Jinwen Wu, Zijun Lu, Muhammad Qasim Shahid. Autotetraploid rice: challenges and opportunities [J]. Hereditas(Beijing), 2023, 45(9): 781-792. |
| [6] | Wenrui Shi, Hongzhu Qu, Xiangdong Fang. Overview of multi-omics research in gout [J]. Hereditas(Beijing), 2023, 45(8): 643-657. |
| [7] | Fang Wang, Yuebo Zhang, Qian Jiang, Yulong Yin, Bi’e Tan, Jiashun Chen. Analysis of transcriptome differences between subcutaneous and intramuscular adipose tissue of Ningxiang pigs [J]. Hereditas(Beijing), 2023, 45(12): 1147-1157. |
| [8] | Xiufang Ou, Ying Wu, Ning Li, Lili Jiang, Bao Liu, Lei Gong. Epigenetics comprehensive experimental course based on the integration of science and education to cultivate students' ability of cutting-edge innovation [J]. Hereditas(Beijing), 2023, 45(12): 1158-1168. |
| [9] | Yanan Li, Xianjun Zhang, Ning Zhang, Yalin Liang, Yuxing Zhang, Huaxing Zhao, Zicong Li, Sixiu Huang. Effects of overexpression of histone H3K9me3 demethylase on development of porcine cloned embryos [J]. Hereditas(Beijing), 2023, 45(1): 67-77. |
| [10] | Fei Gao, Yu Wang, Jiaxiang Du, Xuguang Du, Jianguo Zhao, Dengke Pan, Sen Wu, Yaofeng Zhao. Advances and applications of genetically modified pig models in biomedical and agricultural field [J]. Hereditas(Beijing), 2023, 45(1): 6-28. |
| [11] | Mengxuan Xu, Ming Zhou. Advances of RNA polymerase IV in controlling DNA methylation and development in plants [J]. Hereditas(Beijing), 2022, 44(7): 567-580. |
| [12] | Yan Zhao, Chenxin Wang, Tianming Yang, Chunshuang Li, Lihong Zhang, Dongni Du, Ruoxi Wang, Jing Wang, Min Wei, Xueqing Ba. Linking oxidative DNA lesion 8-OxoG to tumor development and progression [J]. Hereditas(Beijing), 2022, 44(6): 466-477. |
| [13] | Hui Qu, Yi Liu, Yawen Chen, Hui Wang. Alteration of imprinted genes and offspring organ development caused by environmental factors [J]. Hereditas(Beijing), 2022, 44(2): 107-116. |
| [14] | Ziwen Shi, Qing He, Zhuofan Zhao, Xiaowei Liu, Peng Zhang, Moju Cao. Exploration and utilization of maize male sterility resources [J]. Hereditas(Beijing), 2022, 44(2): 134-152. |
| [15] | Yangjinghui Zhang, Peiyao Chang, Zishu Yang, Yuhang Xue, Xueqi Li, Yang Zhang. Advances in epigenetic modification affecting anthocyanin synthesis [J]. Hereditas(Beijing), 2022, 44(12): 1117-1127. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||