Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (6): 466-477.doi: 10.16288/j.yczz.22-067
• Review • Previous Articles Next Articles
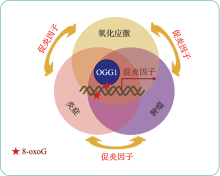
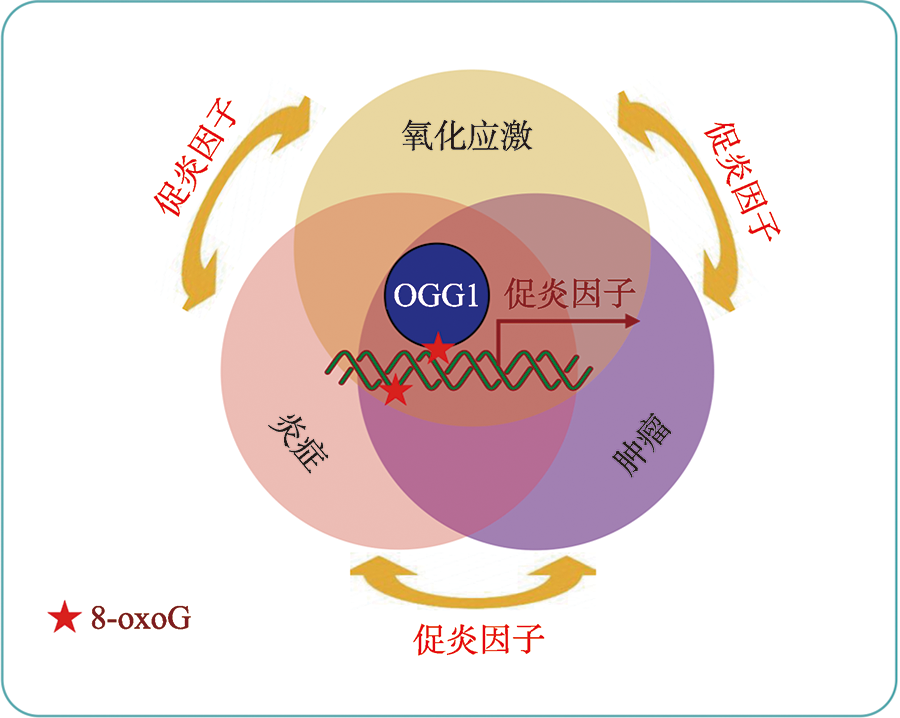
Linking oxidative DNA lesion 8-OxoG to tumor development and progression
Yan Zhao1, Chenxin Wang2( ), Tianming Yang2, Chunshuang Li2, Lihong Zhang2, Dongni Du2, Ruoxi Wang3, Jing Wang4, Min Wei2, Xueqing Ba2(
), Tianming Yang2, Chunshuang Li2, Lihong Zhang2, Dongni Du2, Ruoxi Wang3, Jing Wang4, Min Wei2, Xueqing Ba2( )
)
- 1. Department of Biological Vector Control, Center for Disease Control and Prevention of Jilin Province, Changchun 130062, China
2. The Key Laboratory of Molecular Epigenetics of Ministry of Education, College of Life Sciences, Northeast Normal University, Changchun 130024, China
3. College of Life Sciences, Shandong Normal University, Jinan 250014, China
4. Department of Respiratory Medicine, China-Japan Union Hospital of Jilin University, Changchun 130061, China
-
Received:2022-03-10Revised:2022-04-16Online:2022-06-20Published:2022-04-22 -
Contact:Ba Xueqing E-mail:wangcx968@nenu.edu.cn;baxq755@nenu.edu.cn -
Supported by:Nos.Supported by the National Natural Science Foundation of China(31900557);Nos.FSupported by the National Natural Science Foundation of China(32170591);Nos.Supported by the National Natural Science Foundation of China(Nos.31970686);the Natural Science Foundation of Jilin Province of China(No. 20210101356JC)
Cite this article
Yan Zhao, Chenxin Wang, Tianming Yang, Chunshuang Li, Lihong Zhang, Dongni Du, Ruoxi Wang, Jing Wang, Min Wei, Xueqing Ba. Linking oxidative DNA lesion 8-OxoG to tumor development and progression[J]. Hereditas(Beijing), 2022, 44(6): 466-477.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Radak Z, Boldogh I. 8-Oxo-7,8-dihydroguanine: links to gene expression, aging, and defense against oxidative stress. Free Radic Biol Med, 2010, 49(4):587-596.
doi: 10.1016/j.freeradbiomed.2010.05.008 |
| [2] |
Friedberg EC. DNA damage and repair. Nature, 2003, 421(6921):436-440.
doi: 10.1038/nature01408 |
| [3] |
Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008, 18(1):27-47.
doi: 10.1038/cr.2008.8 |
| [4] | Hegde ML, Izumi T, Mitra S. Oxidized base damage and single-strand break repair in mammalian genomes: role of disordered regions and posttranslational modifications in early enzymes. Prog Mol Biol Transl Sci, 2012, 110:123-153. |
| [5] |
Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem Rev, 1998, 98(3):1109-1152.
doi: 10.1021/cr960421s |
| [6] | Candeias LP, Steenken S. Reaction of HO* with guanine derivatives in aqueous solution: formation of two different redox-active OH-adduct radicals and their unimolecular transformation reactions. Properties of G(-H)*. Chemistry, 2000, 6(3):475-484. |
| [7] |
Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet, 1993, 9(7):246-249.
pmid: 8379000 |
| [8] |
Hart RW, Setlow RB. Correlation between deoxyribonucleic acid excision-repair and life-span in a number of mammalian species. Proc Natl Acad Sci USA, 1974, 71(6):2169-2173.
doi: 10.1073/pnas.71.6.2169 |
| [9] |
Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci USA, 1993, 90(17):7915-7922.
doi: 10.1073/pnas.90.17.7915 |
| [10] |
Ba XQ, Aguilera-Aguirre L, Sur S, Boldogh I. 8-Oxoguanine DNA glycosylase-1-driven DNA base excision repair: role in asthma pathogenesis. Curr Opin Allergy Clin Immunol, 2015, 15(1):89-97.
doi: 10.1097/ACI.0000000000000135 |
| [11] |
Ba XQ, Bacsi A, Luo JX, Aguilera-Aguirre L, Zeng XL, Radak Z, Brasier AR, Boldogh I. 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J Immunol, 2014, 192(5):2384-2394.
doi: 10.4049/jimmunol.1302472 |
| [12] |
Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci USA, 1974, 71(9):3649-3653.
doi: 10.1073/pnas.71.9.3649 |
| [13] | Krokan HE, Bjørås M. Base excision repair. Cold Spring Harb Perspect Biol, 2013, 5(4):a012583. |
| [14] |
Doetsch PW, Cunningham RP. The enzymology of apurinic/apyrimidinic endonucleases. Mutat Res, 1990, 236(2-3):173-201.
doi: 10.1016/0921-8777(90)90004-O |
| [15] |
Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science, 1995, 269(5224):699-702.
doi: 10.1126/science.7624801 pmid: 7624801 |
| [16] |
Ba XQ, Aguilera-Aguirre L, Rashid QTAN, Bacsi A, Radak Z, Sur S, Hosoki K, Hegde ML, Boldogh I. The role of 8-oxoguanine DNA glycosylase-1 in inflammation. Int J Mol Sci, 2014, 15(9):16975-16997.
doi: 10.3390/ijms150916975 |
| [17] |
David SS, O'Shea VL,Kundu S. Base-excision repair of oxidative DNA damage. Nature, 2007, 447(7147):941-950.
doi: 10.1038/nature05978 |
| [18] |
Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic Biol Med, 2008, 45(12):1610-1621.
doi: 10.1016/j.freeradbiomed.2008.07.004 |
| [19] |
Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature, 2000, 403(6772):859-866.
doi: 10.1038/35002510 |
| [20] |
Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology, 2003, 193(1-2):43-65.
doi: 10.1016/S0300-483X(03)00289-0 |
| [21] |
Nakabeppu Y. Cellular levels of 8-oxoguanine in either DNA or the nucleotide pool play pivotal roles in carcinogenesis and survival of cancer cells. Int J Mol Sci, 2014, 15(7):12543-12557.
doi: 10.3390/ijms150712543 pmid: 25029543 |
| [22] |
Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol, 1992, 174(20):6321-6325.
pmid: 1328155 |
| [23] |
Bos JL. Ras oncogenes in human cancer: a review. Cancer Res, 1989, 49(17):4682-4689.
pmid: 2547513 |
| [24] |
Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol Mol Biol Rev, 2004, 68(2):320-344.
doi: 10.1128/MMBR.68.2.320-344.2004 |
| [25] |
Khosravi-Far R, Der CJ. The Ras signal transduction pathway. Cancer Metastasis Rev, 1994, 13(1):67-89.
doi: 10.1007/BF00690419 |
| [26] |
Mills NE, Fishman CL, Rom WN, Dubin N, Jacobson DR. Increased prevalence of K-ras oncogene mutations in lung adenocarcinoma. Cancer Res, 1995, 55(7):1444-1447.
pmid: 7882350 |
| [27] |
Sugio K, Ishida T, Yokoyama H, Inoue T, Sugimachi K, Sasazuki T. Ras gene mutations as a prognostic marker in adenocarcinoma of the human lung without lymph node metastasis. Cancer Res, 1992, 52(10):2903-2906.
pmid: 1581907 |
| [28] |
Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature, 2001, 410(6832):1111-1116.
doi: 10.1038/35074129 |
| [29] |
Osterod M, Hollenbach S, Hengstler JG, Barnes DE, Lindahl T, Epe B. Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8- oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis, 2001, 22(9):1459-1463.
pmid: 11532868 |
| [30] |
Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA, 1999, 96(23):13300-13305.
doi: 10.1073/pnas.96.23.13300 |
| [31] |
Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, Itoh M, Takano H, Hippou Y, Aburatani H, Masumura K, Nohmi T, Nishimura S, Noda T. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci USA, 2000, 97(8):4156-4161.
doi: 10.1073/pnas.050404497 |
| [32] |
Arai T, Kelly VP, Minowa O, Noda T, Nishimura S. High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis, 2002, 23(12):2005-2010.
doi: 10.1093/carcin/23.12.2005 |
| [33] | Sakumi K, Tominaga Y, Furuichi M, Xu P, Tsuzuki T, Sekiguchi M, Nakabeppu Y. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res, 2003, 63(5):902-905. |
| [34] |
Al-Tassan N, Chmiel NH, Maynard J, Fleming N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS, Sampson JR, Cheadle JP. Inherited variants of MYH associated with somatic G:C-->T:A mutations in colorectal tumors. Nat Genet, 2002, 30(2):227-232.
pmid: 11818965 |
| [35] |
Sieber OM, Lipton L, Crabtree M, Heinimann K, Fidalgo P, Phillips RKS, Bisgaard M-L, Orntoft TF, Aaltonen LA, Hodgson SV, Thomas HJW, Tomlinson IPM. Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N Engl J Med, 2003, 348(9):791-799.
doi: 10.1056/NEJMoa025283 |
| [36] |
D'Onofrio G, Jabbari K,Musto H,Alvarez-Valin F,Cruveiller S,Bernardi G. Evolutionary genomics of vertebrates and its implications. Ann N Y Acad Sci, 1999, 870:81-94.
doi: 10.1111/j.1749-6632.1999.tb08867.x |
| [37] |
Gautier C. Compositional bias in DNA. Curr Opin Genet Dev, 2000, 10(6):656-661.
pmid: 11088017 |
| [38] |
Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA, 2006, 103(5):1412-1417.
doi: 10.1073/pnas.0510310103 |
| [39] |
Hall DB, Holmlin RE, Barton JK. Oxidative DNA damage through long-range electron transfer. Nature, 1996, 382(6593):731-735.
doi: 10.1038/382731a0 |
| [40] |
Giese B, Amaudrut J, Köhler AK, Spormann M, Wessely S. Direct observation of hole transfer through DNA by hopping between adenine bases and by tunnelling. Nature, 2001, 412(6844):318-320.
doi: 10.1038/35085542 |
| [41] |
Núñez ME, Hall DB, Barton JK. Long-range oxidative damage to DNA: effects of distance and sequence. Chem Biol, 1999, 6(2):85-97.
doi: 10.1016/S1074-5521(99)80005-2 |
| [42] | Saito I, Takayama M, Nakamura T, Sugiyama H, Komeda Y, Iwasaki M. The most electron-donating sites in duplex DNA: guanine-guanine stacking rule. Nucleic Acids Symp Ser, 1995, (34):191-192. |
| [43] |
Genereux JC, Boal AK, Barton JK. DNA-mediated charge transport in redox sensing and signaling. J Am Chem Soc, 2010, 132(3):891-905.
doi: 10.1021/ja907669c pmid: 20047321 |
| [44] |
Merino EJ, Boal AK, Barton JK. Biological contexts for DNA charge transport chemistry. Curr Opin Chem Biol, 2008, 12(2):229-237.
doi: 10.1016/j.cbpa.2008.01.046 |
| [45] |
Giese B. Long-distance electron transfer through DNA. Annu Rev Biochem, 2002, 71:51-70.
doi: 10.1146/annurev.biochem.71.083101.134037 |
| [46] |
Ohno M, Miura T, Furuichi M, Tominaga Y, Tsuchimoto D, Sakumi K, Nakabeppu Y. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res, 2006, 16(5):567-575.
doi: 10.1101/gr.4769606 |
| [47] |
Ding Y, Fleming AM, Burrows CJ. Sequencing the mouse genome for the oxidatively modified base 8-oxo-7,8- dihydroguanine by OG-Seq. J Am Chem Soc, 2017, 139(7):2569-2572.
doi: 10.1021/jacs.6b12604 pmid: 28150947 |
| [48] |
Pastukh V, Roberts JT, Clark DW, Bardwell GC, Patel M, Al-Mehdi AB, Borchert GM, Gillespie MN. An oxidative DNA "damage" and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am J Physiol Lung Cell Mol Physiol, 2015, 309(11):L1367-1375.
doi: 10.1152/ajplung.00236.2015 |
| [49] |
Yoshihara M, Jiang L, Akatsuka S, Suyama M, Toyokuni S. Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res, 2014, 21(6):603-612.
doi: 10.1093/dnares/dsu023 pmid: 25008760 |
| [50] |
Kawai K, Wata Y, Hara M, Tojo S, Majima T. Regulation of one-electron oxidation rate of guanine by base pairing with cytosine derivatives. J Am Chem Soc, 2002, 124(14):3586-3590.
doi: 10.1021/ja016530s |
| [51] |
Ming X, Matter B, Song M, Veliath E, Shanley R, Jones R, Tretyakova N. Mapping structurally defined guanine oxidation products along DNA duplexes: influence of local sequence context and endogenous cytosine methylation. J Am Chem Soc, 2014, 136(11):4223-4235.
doi: 10.1021/ja411636j pmid: 24571128 |
| [52] |
Wang RX, Hao WJ, Pan L, Boldogh I, Ba XQ. The roles of base excision repair enzyme OGG1 in gene expression. Cell Mol Life Sci, 2018, 75(20):3741-3750.
doi: 10.1007/s00018-018-2887-8 |
| [53] |
Hao WJ, Qi TY, Pan L, Wang RX, Zhu B, Aguilera- Aguirre L, Radak Z, Hazra TK, Vlahopoulos SA, Bacsi A, Brasier AR, Ba XQ, Boldogh I. Effects of the stimuli- dependent enrichment of 8-oxoguanine DNA glycosylase1 on chromatinized DNA. Redox Biol, 2018, 18:43-53.
doi: 10.1016/j.redox.2018.06.002 |
| [54] |
Hänsel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol, 2017, 18(5):279-284.
doi: 10.1038/nrm.2017.3 |
| [55] |
Chio IIC, Tuveson DA. ROS in cancer: the burning question. Trends Mol Med, 2017, 23(5):411-429.
doi: 10.1016/j.molmed.2017.03.004 |
| [56] |
Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol, 2009, 19(8):414-422.
doi: 10.1016/j.tcb.2009.05.002 pmid: 19589679 |
| [57] |
Fleming AM, Zhu J, Ding Y, Visser JA, Zhu J, Burrows CJ. Human DNA repair genes possess potential G-quadruplex sequences in their promoters and 5'-untranslated regions. Biochemistry, 2018, 57(6):991-1002.
doi: 10.1021/acs.biochem.7b01172 pmid: 29320161 |
| [58] |
Fleming AM, Zhu J, Ding Y, Burrows CJ. 8-Oxo-7,8- dihydroguanine in the Context of a Gene Promoter G-Quadruplex Is an On-Off Switch for Transcription. ACS Chem Biol, 2017, 12(9):2417-2426.
doi: 10.1021/acschembio.7b00636 pmid: 28829124 |
| [59] |
Fleming AM, Ding Y, Burrows CJ. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc Natl Acad Sci USA, 2017, 114(10):2604-2609.
doi: 10.1073/pnas.1619809114 |
| [60] |
Kitsera N, Stathis D, Lühnsdorf B, Müller H, Carell T, Epe B, Khobta A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res, 2011, 39(14):5926-5934.
doi: 10.1093/nar/gkr163 |
| [61] |
An J, Yin MD, Yin JY, Wu SZ, Selby CP, Yang YY, Sancar A, Xu G-L, Qian MX, Hu JC. Genome-wide analysis of 8-oxo-7,8-dihydro-2'-deoxyguanosine at single-nucleotide resolution unveils reduced occurrence of oxidative damage at G-quadruplex sites. Nucleic Acids Res, 2021, 49(21):12252-12267.
doi: 10.1093/nar/gkab1022 |
| [62] |
Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res, 2006, 34(9):2536-2549.
doi: 10.1093/nar/gkl286 |
| [63] |
Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA, 2002, 99(18):11593-11598.
doi: 10.1073/pnas.182256799 |
| [64] |
Cogoi S, Ferino A, Miglietta G, Pedersen EB, Xodo LE. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res, 2018, 46(2):661-676.
doi: 10.1093/nar/gkx1142 |
| [65] |
Ruchko MV, Gorodnya OM, Pastukh VM, Swiger BM, Middleton NS, Wilson GL, Gillespie MN. Hypoxia-induced oxidative base modifications in the VEGF hypoxia- response element are associated with transcriptionally active nucleosomes. Free Radic Biol Med, 2009, 46(3):352-359.
doi: 10.1016/j.freeradbiomed.2008.09.038 |
| [66] |
Xia LN, Huang WJ, Bellani M, Seidman MM, Wu KC, Fan DM, Nie YZ, Cai Y, Zhang YW, Yu L-R, Li HL, Zahnow CA, Xie WB, Chiu Yen R-W, Rassool FV, Baylin SB. CHD4 Has Oncogenic Functions in Initiating and Maintaining Epigenetic Suppression of Multiple Tumor Suppressor Genes. Cancer Cell, 2017, 31(5): 653-668.e657.
doi: 10.1016/j.ccell.2017.04.005 |
| [67] |
Zhou XL, Wang WT, Du CT, Yan FF, Yang SB, He K, Wang H, Zhao AY. OGG1 regulates the level of symmetric dimethylation of histone H4 arginine-3 by interacting with PRMT5. Mol Cell Probes, 2018, 38:19-24.
doi: 10.1016/j.mcp.2018.01.002 |
| [68] |
Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science, 2008, 319(5860):202-206.
doi: 10.1126/science.1147674 pmid: 18187655 |
| [69] | Zhu F, Rui LX. PRMT5 in gene regulation and hematologic malignancies. Genes Dis, 2019, 6(3):247-257. |
| [70] | Motolani A, Martin M, Sun MY, Lu T. The Structure and Functions of PRMT5 in Human Diseases. Life (Basel), 2021, 11(10):1074. |
| [71] |
Touati E, Michel V, Thiberge J-M, Avé P, Huerre M, Bourgade F, Klungland A, Labigne A. Deficiency in OGG1 protects against inflammation and mutagenic effects associated with H. pylori infection in mouse. Helicobacter, 2006, 11(5):494-505.
doi: 10.1111/j.1523-5378.2006.00442.x |
| [72] |
Mabley JG, Pacher P, Deb A, Wallace R, Elder RH, Szabo C. Potential role for 8-oxoguanine DNA glycosylase in regulating inflammation. FASEB J, 2005, 19(2):290-292.
pmid: 15677345 |
| [73] |
Li GQ, Yuan KF, Yan CG, Fox Jr, Gaid M, Breitwieser W, Bansal AK, Zeng HW, Gao HW, Wu M. 8-Oxoguanine- DNA glycosylase 1 deficiency modifies allergic airway inflammation by regulating STAT6 and IL-4 in cells and in mice. Free Radic Biol Med, 2012, 52(2):392-401.
doi: 10.1016/j.freeradbiomed.2011.10.490 |
| [74] |
Pan L, Zhu B, Hao WJ, Zeng XL, Vlahopoulos SA, Hazra TK, Hegde ML, Radak Z, Bacsi A, Brasier AR, Ba XQ, Boldogh I. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor kappaB-driven gene expression. J Biol Chem, 2016, 291(49):25553-25566.
doi: 10.1074/jbc.M116.751453 |
| [75] |
Pan L, Hao WJ, Zheng X, Zeng XL, Ahmed Abbasi A, Boldogh I, Ba XQ. OGG1-DNA interactions facilitate NF-kappaB binding to DNA targets. Sci Rep, 2017, 7:43297.
doi: 10.1038/srep43297 |
| [76] |
Hao WJ, Wang J, Zhang YH, Wang CX, Xia L, Zhang WH, Zafar M, Kang J-Y, Wang RX, Ali Bohio A, Pan L, Zeng XL, Wei M, Boldogh I, Ba XQ. Enzymatically inactive OGG1 binds to DNA and steers base excision repair toward gene transcription. FASEB J, 2020, 34(6):7427-7441.
doi: 10.1096/fj.201902243R |
| [77] |
Visnes T, Cázares-Körner A, Hao WJ, Wallner O, Masuyer G, Loseva O, Mortusewicz O, Wiita E, Sarno A, Manoilov A, Astorga-Wells J, Jemth A-S, Pan L, Sanjiv K, Karsten S, Gokturk C, Grube M, Homan EJ, Hanna BMF, Paulin CBJ, Pham T, Rasti A, Berglund UW,von Nicolai C,Benitez-Buelga C,Koolmeister T,Ivanic D,Iliev P,Scobie M,Krokan HE,Baranczewski P,Artursson P,Altun M,Jensen AJ,Kalderén C,Ba XQ,Zubarev RA,Stenmark P,Boldogh I,Helleday T,. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science, 2018, 362(6416):834-839.
doi: 10.1126/science.aar8048 |
| [78] |
Sen CK, Packer L. Antioxidant and redox regulation of gene transcription. FASEB J, 1996, 10(7):709-720.
pmid: 8635688 |
| [79] | Cheung EC, Vousden KH. The role of ROS in tumour development and progression. Nat Rev Cancer, 2022. |
| [80] |
Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell, 2005, 121(5):667-670.
doi: 10.1016/j.cell.2005.05.016 |
| [81] |
Propper DJ, Balkwill FR. Harnessing cytokines and chemokines for cancer therapy. Nat Rev Clin Oncol, 2022, 19(4):237-253.
doi: 10.1038/s41571-021-00588-9 pmid: 34997230 |
| [82] |
Heath O, Berlato C, Maniati E, Lakhani A, Pegrum C, Kotantaki P, Elorbany S, Bohm S, Barry ST, Annibaldi A, Barton DP, Balkwill FR. Chemotherapy induces tumor- associated macrophages that aid adaptive immune responses in ovarian cancer. Cancer Immunol Res, 2021, 9(6):665-681.
doi: 10.1158/2326-6066.CIR-20-0968 pmid: 33839687 |
| [83] |
Shi HF, Han XQ, Sun YY, Shang C, Wei M, Ba XQ, Zeng XL. Chemokine (C-X-C motif) ligand 1 and CXCL2 produced by tumor promote the generation of monocytic myeloid-derived suppressor cells. Cancer Sci, 2018, 109(12):3826-3839.
doi: 10.1111/cas.13809 |
| [84] |
Han XQ, Shi HF, Sun YY, Shang C, Luan T, Wang DK, Ba XQ, Zeng XL. CXCR2 expression on granulocyte and macrophage progenitors under tumor conditions contributes to mo-MDSC generation via SAP18/ERK/STAT3. Cell Death Dis, 2019, 10(8):598.
doi: 10.1038/s41419-019-1837-1 |
| [85] |
Ohl K, Tenbrock K. Reactive oxygen species as regulators of MDSC-mediated immune suppression. Front Immunol, 2018, 9:2499.
doi: 10.3389/fimmu.2018.02499 |
| [86] |
Seifermann M, Epe B. Oxidatively generated base modifications in DNA: Not only carcinogenic risk factor but also regulatory mark? Free Radic Biol Med, 2017, 107:258-265.
doi: 10.1016/j.freeradbiomed.2016.11.018 |
| [87] |
Ba XQ, Boldogh I. 8-Oxoguanine DNA glycosylase 1: Beyond repair of the oxidatively modified base lesions. Redox Biol, 2018, 14:669-678.
doi: 10.1016/j.redox.2017.11.008 |
| [88] | Yu C, Dong CR, Zhang ZH, Li Q, Zeng HH. Review of the research on 8-hydroxy-2 deoxyguanosine as a DNA oxidative damage marker. Chin J Clin Pharmacol, 2017, 33(13):1267-1270. |
| [1] | Qingyu Sun, Yang Zhou, Lijuan Du, Mengke Zhang, Jiale Wang, Yuanyuan Ren, Fang Liu. Analysis between macrophage-related genes with prognosis and tumor microenvironment in non-small cell lung cancer [J]. Hereditas(Beijing), 2023, 45(8): 684-699. |
| [2] | Chenghao Yan, Weiyu Bai, Zhimeng Zhang, Junling Shen, Youjun Wang, Jianwei Sun. The roles and mechanism of STIM1 in tumorigenesis and metastasis [J]. Hereditas(Beijing), 2023, 45(5): 395-408. |
| [3] | Shunze Wang, Feng Jiang, Dongli Zhu, Tie-Lin Yang, Yan Guo. Application of Hi-C technology in three-dimensional genomics research and disease pathogenesis analysis [J]. Hereditas(Beijing), 2023, 45(4): 279-294. |
| [4] | Chunhui Ma, Haixu Hu, Lijuan Zhang, Yi Liu, Tianyi Liu. Establishment and verification of a digital PCR assay for the detection of CK19 expression in quantitative analysis of circulating tumor cell [J]. Hereditas(Beijing), 2023, 45(3): 250-260. |
| [5] | Dong Chang, Xiangxiang Liu, Rui Liu, Jianwei Sun. The role and regulatory mechanism of FSCN1 in breast tumorigenesis and progression [J]. Hereditas(Beijing), 2023, 45(2): 115-127. |
| [6] | Qinggang Hao, Fenggui Sun, Chenghao Yan, Jianwei Sun. Progress on the role and mechanism of MT1-MMP in tumor metastasis [J]. Hereditas(Beijing), 2022, 44(9): 745-755. |
| [7] | Siyuan Xu, Jia Shou, Qiang Wu. Additional evidence of HS5-1 enhancer eRNA PEARL for protocadherin alpha gene regulation [J]. Hereditas(Beijing), 2022, 44(8): 695-764. |
| [8] | Qianbin Zhu, Zhicheng Gan, Xiaocui Li, Yingjie Zhang, Heming Zhao, Xianzhong Huang. Genome-wide identification, phylogenetic and expression of MAPKKK gene family in Arabidopsis pumila [J]. Hereditas(Beijing), 2022, 44(11): 1044-1055. |
| [9] | Jiangping He, Jiekai Chen. Epigenetic control of transposable elements and cell fate decision [J]. Hereditas(Beijing), 2021, 43(9): 822-834. |
| [10] | Cong Zhou, Qiangwei Zhou, Sheng Cheng, Guoliang Li. Research progress of CTCF in mediating 3D genome formation and regulating gene expression [J]. Hereditas(Beijing), 2021, 43(9): 816-821. |
| [11] | Yanni Kou, Shan Cen, Xiaoyu Li. Research and application on LINE-1 in diagnosis and treatment of tumorigenesis [J]. Hereditas(Beijing), 2021, 43(6): 571-579. |
| [12] |
Zhuo Wang, Xiaohan Shen, Qihui Shi.
|
| [13] | Haidong Xu, Bolin Ning, Fang Mu, Hui Li, Ning Wang. Advances of functional consequences and regulation mechanisms of alternative cleavage and polyadenylation [J]. Hereditas(Beijing), 2021, 43(1): 4-15. |
| [14] | Linan Zhao, Na Wang, Guoliang Yang, Xianbin Su, Zeguang Han. A method for reliable detection of genomic point mutations based on single-cell target-sequencing [J]. Hereditas(Beijing), 2020, 42(7): 703-712. |
| [15] | Shumin Chen, Ling Ma, Shan Cen. Progress of SLFN family proteins in tumor and virus infection [J]. Hereditas(Beijing), 2020, 42(5): 444-451. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||