Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (10): 860-870.doi: 10.16288/j.yczz.24-221
• Research Article • Previous Articles Next Articles
Screening and analysis of GULP1 downstream target genes based on transcriptomic sequencing
Xin Wen( ), Jin Mei, Meiyu Qian, Yidan Jiang, Juan Wang, Shibo Xu, Cuizhe Wang, Jun Zhang(
), Jin Mei, Meiyu Qian, Yidan Jiang, Juan Wang, Shibo Xu, Cuizhe Wang, Jun Zhang( )
)
- Key laboratory of Xinjiang Endemic and Ethnic Diseases, School of Medicine,Shihezi University, Shihezi 832000, China
-
Received:2024-08-02Revised:2024-09-01Online:2024-09-02Published:2024-09-02 -
Contact:Jun Zhang E-mail:1391476108@qq.com;zhangjunyc@.163.com -
Supported by:National Natural Science Foundation of China Project(82260162);Xinjiang Uyghur Autonomous Region “Tianshan Talent” Project(2023TSYCCX0116);Xinjiang Uyghur Autonomous Region “Tianshan Talent” Project(2023TSYCQNTJ0032);Xinjiang Production and Construction Corps Guided Science and Technology Plan Project(2022ZD001);Xinjiang Production and Construction Corps Guided Science and Technology Plan Project(2023ZD037)
Cite this article
Xin Wen, Jin Mei, Meiyu Qian, Yidan Jiang, Juan Wang, Shibo Xu, Cuizhe Wang, Jun Zhang. Screening and analysis of GULP1 downstream target genes based on transcriptomic sequencing[J]. Hereditas(Beijing), 2024, 46(10): 860-870.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
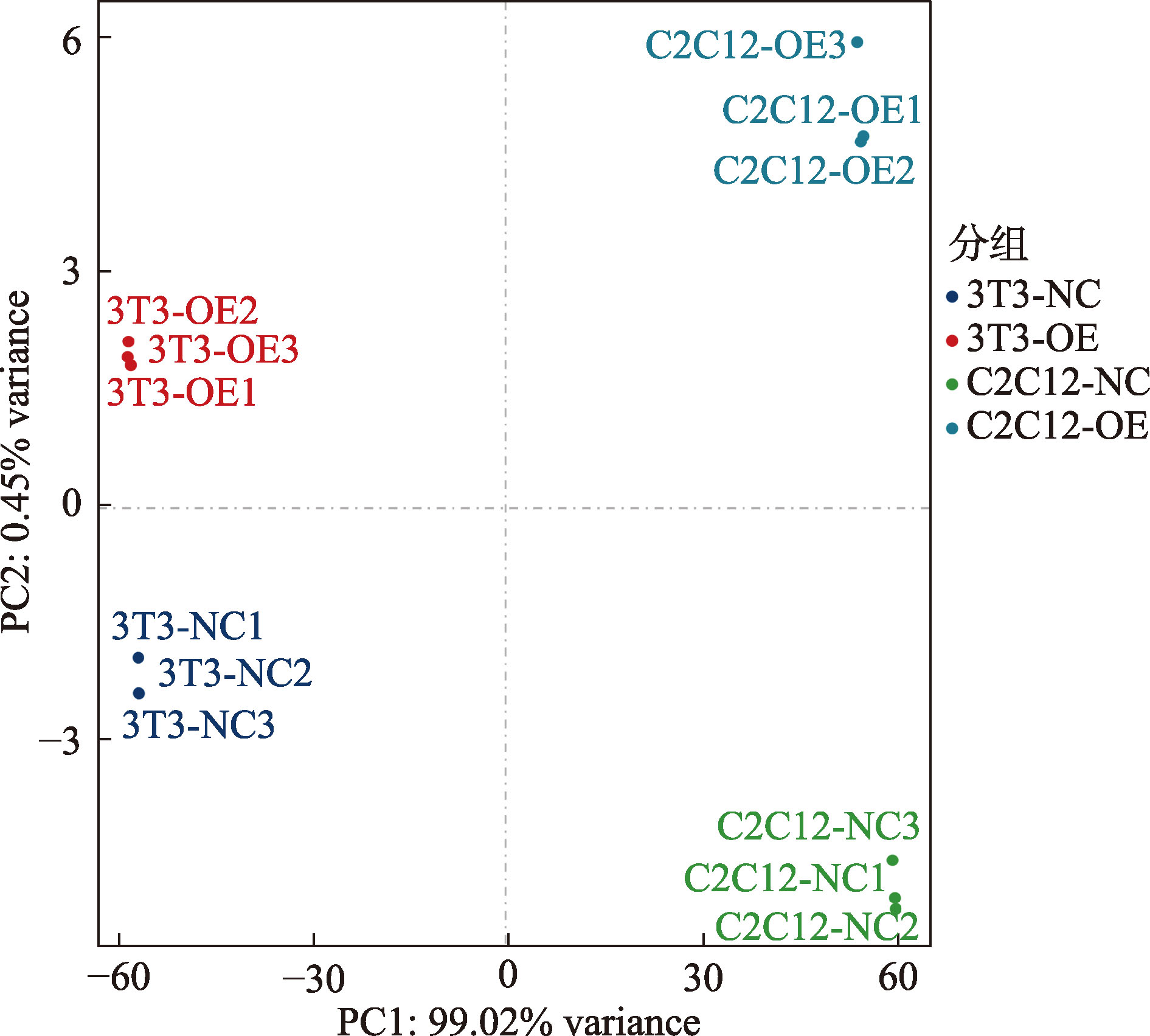
Table 1
Statistics of sequence data after filtering"
| 样本 | 原始 reads 数目 | 原始 测序量 | 过滤后的Reads数目 | 过滤后 的测序量 | 有效碱基 百分比 | Q30(%) | GC(%) |
|---|---|---|---|---|---|---|---|
| 3T3-NC1 | 48.75 | 7.18 | 47.51 | 7 | 97.45 | 95.13 | 51.88 |
| 3T3-NC2 | 48.84 | 7.18 | 47.45 | 6.97 | 97.15 | 95.18 | 52.55 |
| 3T3-NC3 | 47.86 | 7.06 | 46.7 | 6.89 | 97.57 | 95.11 | 52.56 |
| 3T3-OE1 | 47.87 | 7.06 | 46.71 | 6.89 | 97.56 | 95.19 | 52.25 |
| 3T3-OE2 | 49.11 | 7.2 | 47.63 | 6.99 | 96.99 | 95.08 | 51.98 |
| 3T3-OE3 | 48.65 | 7.18 | 47.49 | 7.01 | 97.6 | 95.34 | 52.52 |
| C2C12-NC1 | 48.39 | 7.12 | 47.06 | 6.93 | 97.26 | 95.17 | 52.9 |
| C2C12-NC2 | 48.7 | 7.2 | 47.52 | 7.02 | 97.56 | 95.1 | 52.87 |
| C2C12-NC3 | 48.5 | 7.12 | 47.11 | 6.92 | 97.13 | 95.23 | 52.53 |
| C2C12-OE1 | 48.42 | 7.1 | 46.95 | 6.89 | 96.96 | 95.08 | 52.42 |
| C2C12-OE2 | 48.21 | 7.1 | 46.91 | 6.91 | 97.3 | 95.11 | 53.01 |
| C2C12-OE3 | 48.32 | 7.14 | 47.25 | 6.99 | 97.79 | 95.05 | 52.3 |
Table 2
PCR primer sequences of the target genes"
| 基因 | 上游引物序列(5′→3′) | 下游引物序列(5′→3′) |
|---|---|---|
| Abca1 | GCTTGTTGGCCTCAGTTAAGG | GTAGCTCAGGCGTACAGAGAT |
| Ccl3 | TTCTCTGTACCATGACACTCTGC | CGTGGAATCTTCCGGCTGTAG |
| Ccl5 | GCTGCTTTGCCTACCTCTCC | TCGAGTGACAAACACGACTGC |
| Fos | CGGGTTTCAACGCCGACTA | TTGGCACTAGAGACGGACAGA |
| Il6 | TAGTCCTTCCTACCCCAATTTCC | TTGGTCCTTAGCCACTCCTTC |
| Lbp | GATCACCGACAAGGGCCTG | GGCTATGAAACTCGTACTGCC |
| Map2k6 | ATGTCTCAGTCGAAAGGCAAG | TTGGAGTCTAAATCCCGAGGC |
| Map3k8 | ATGGAGTACATGAGCACTGGA | GGCTCTTCACTTGCATAAAGGTT |
| Tlr7 | ATGTGGACACGGAAGAGACAA | GGTAAGGGTAAGATTGGTGGTG |
| Angpt4 | CAGCCAGCTATGCTACTAGATGG | CCTCTGGAGGCTATTGGAGC |
| Gngt2 | CAGGACCTCAGTGAGAAGGAG | CCTGCTTGGGCCTCTACATAAT |
| Il2rb | TGGAGCCTGTCCCTCTACG | TCCACATGCAAGAGACATTGG |
| Ret | TTTCTCAAGGGATGCTTACTGGG | CCCGTAGGGCATGGACATAGA |
| Adora2a | GCCATCCCATTCGCCATCA | GCAATAGCCAAGAGGCTGAAGA |
| Atp1b2 | GGCAGGTGGTTGAGGAGTG | GGGGTATGGTCAGAGACGGT |
| Calml4 | TCTTCTGGTGTCCATGAGGTG | GAAGTCCAGCTCTCCGTTCTT |
| Npr1 | GCTTGTGCTCTATGCAGATCG | TCGACGAACTCCTGGTGATTTA |
| Npy1r | GCTTGTGCTCTATGCAGATCG | TCGACGAACTCCTGGTGATTTA |
| [1] |
Sharifi S, Daghighi S, Motazacker MM, Badlou B, Sanjabi B, Akbarkhanzadeh A, Rowshani AT, Laurent S, Peppelenbosch MP, Rezaee F. Superparamagnetic iron oxide nanoparticles alter expression of obesity and T2D-associated risk genes in human adipocytes. Sci Rep, 2013, 3: 2173.
doi: 10.1038/srep02173 pmid: 23838847 |
| [2] |
Kiss RS, Ma Z, Nakada-Tsukui K, Brugnera E, Vassiliou G, McBride HM, Ravichandran KS, Marcel YL. The lipoprotein receptor-related protein-1 (LRP) adapter protein GULP mediates trafficking of the LRP ligand prosaposin, leading to sphingolipid and free cholesterol accumulation in late endosomes and impaired efflux. J Biol Chem, 2006, 281(17): 12081-12092.
doi: 10.1074/jbc.M600621200 pmid: 16497666 |
| [3] |
Lee CY, Ruel I, Denis M, Genest J, Kiss RS. Cholesterol trapping in Niemann-Pick disease type B fibroblasts can be relieved by expressing the phosphotyrosine binding domain of GULP. J Clin Lipidol, 2013, 7(2): 153-164.
doi: 10.1016/j.jacl.2012.02.006 pmid: 23415435 |
| [4] |
Trommsdorff M, Borg JP, Margolis B, Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem, 1998, 273(50): 33556-33560.
doi: 10.1074/jbc.273.50.33556 pmid: 9837937 |
| [5] |
Ji AL, Meyer JM, Cai L, Akinmusire A, de Beer MC, Webb NR, van der Westhuyzen DR. Scavenger receptor SR-BI in macrophage lipid metabolism. Atherosclerosis, 2011, 217(1): 106-112.
doi: 10.1016/j.atherosclerosis.2011.03.017 pmid: 21481393 |
| [6] | Li RM, Oteiza A, Sørensen KK, McCourt P, Olsen R, Smedsrød B, Svistounov D. Role of liver sinusoidal endothelial cells and stabilins in elimination of oxidized low-density lipoproteins. Am J Physiol Gastrointest Liver Physiol, 2011, 300(1): G71-G81. |
| [7] | Ma CIJ, Martin C, Ma Z, Hafiane A, Dai M, Lebrun JJ, Kiss RS. Engulfment protein GULP is regulator of transforming growth factor-β response in ovarian cells. J Biol Chem, 2012, 287(24): 20636-20651. |
| [8] | Park SY, Kim SY, Kang KB, Kim IS. Adaptor protein GULP is involved in stabilin-1-mediated phagocytosis. Biochem Biophys Res Commun, 2010, 398(3): 467-472. |
| [9] |
Su HP, Nakada-Tsukui K, Tosello-Trampont AC, Li YH, Bu GJ, Henson PM, Ravichandran KS. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP). J Biol Chem, 2002, 277(14): 11772-11779.
doi: 10.1074/jbc.M109336200 pmid: 11729193 |
| [10] |
Sun DJ, Guo YY, Tang PY, Li H, Chen LX. Arf6 as a therapeutic target: Structure, mechanism, and inhibitors. Acta Pharm Sin B, 2023, 13(10): 4089-4104.
doi: 10.1016/j.apsb.2023.06.008 pmid: 37799386 |
| [11] |
Ma Z, Nie ZZ, Luo RB, Casanova JE, Ravichandran KS. Regulation of Arf6 and ACAP1 signaling by the PTB-domain-containing adaptor protein GULP. Curr Biol, 2007, 17(8): 722-727.
doi: 10.1016/j.cub.2007.03.014 pmid: 17398097 |
| [12] | Kim SY, Park SY, Kim JE. GULP1 deficiency reduces adipogenesis and glucose uptake via downregulation of PPAR signaling and disturbing of insulin/ERK signaling in 3T3-L1 cells. J Cell Physiol, 2024, 239(2): e31173. |
| [13] | Hayashi M, Guida E, Inokawa Y, Goldberg R, Reis LO, Ooki A, Pilli M, Sadhukhan P, Woo J, Choi W, Izumchenko E, Gonzalez LM, Marchionni L, Zhavoronkov A, Brait M, Bivalacqua T, Baras A, Netto GJ, Koch W, Singh A, Hoque MO. GULP1 regulates the NRF2-KEAP1 signaling axis in urothelial carcinoma. Sci Signal, 2020, 13(645): eaba0443. |
| [14] | Teramoto Y, Jiang GY, Goto T, Mizushima T, Nagata Y, Netto GJ, Miyamoto H. Androgen receptor signaling induces cisplatin resistance via down-regulating GULP1 expression in bladder cancer. Int J Mol Sci, 2021, 22(18): 10030. |
| [15] |
Maldonado L, Brait M, Izumchenko E, Begum S, Chatterjee A, Sen T, Loyo M, Barbosa A, Poeta ML, Makarev E, Zhavoronkov A, Fazio VM, Angioli R, Rabitti C, Ongenaert M, Van Criekinge W, Noordhuis MG, de Graeff P, Wisman GBA, van der Zee AGJ, Hoque MO. Integrated transcriptomic and epigenomic analysis of ovarian cancer reveals epigenetically silenced GULP1. Cancer Lett, 2018, 433: 242-251.
doi: S0304-3835(18)30436-1 pmid: 29964205 |
| [16] |
Huang MY, Long Y, Jin YZ, Ya WT, Meng DD, Qin TZ, Su LZ, Zhou W, Wu JC, Huang CH, Huang Q. Comprehensive analysis of the lncRNA-miRNA-mRNA regulatory network for bladder cancer. Transl Androl Urol, 2021, 10(3): 1286-1301.
doi: 10.21037/tau-21-81 pmid: 33850763 |
| [17] | Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science, 2020, 368(6487): eaaw5473. |
| [18] | Wang K, Ming H, Zuo J, Tian HL, Huang CH. A review of the redox regulation of tumor metabolism. Journal of Sichuan University (Medical Sciences), 2021, 52(1): 57-63. |
| 王魁, 明慧, 左静, 田海隆, 黄灿华. 氧化还原信号调控与肿瘤代谢. 四川大学学报(医学版), 2021, 52(1): 57-63 | |
| [19] |
Wang ZF, Qin J, Zhao JG, Li JH, Li D, Popp M, Popp F, Alakus H, Kong B, Dong QZ, Nelson PJ, Zhao Y, Bruns CJ. Inflammatory IFIT3 renders chemotherapy resistance by regulating post-translational modification of VDAC2 in pancreatic cancer. Theranostics, 2020, 10(16): 7178-7192.
doi: 10.7150/thno.43093 pmid: 32641986 |
| [20] |
Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun, 2011, 12(6): 399-414.
doi: 10.1038/gene.2011.21 pmid: 21490621 |
| [21] | Yan S, Kumari M, Xiao HP, Jacobs C, Kochumon S, Jedrychowski M, Chouchani E, Ahmad R, Rosen ED. IRF3 reduces adipose thermogenesis via ISG15-mediated reprogramming of glycolysis. J Clin Invest, 2021, 131(7): e144888. |
| [22] |
Xu T, Zhu CZ, Chen JM, Song FF, Ren XX, Wang SS, Yi XF, Zhang YW, Zhang WL, Hu Q, Qin H, Liu YJ, Zhang S, Tan Z, Pan ZF, Huang P, Ge MH. ISG15 and ISGylation modulates cancer stem cell-like characteristics in promoting tumor growth of anaplastic thyroid carcinoma. J Exp Clin Cancer Res, 2023, 42(1): 182.
doi: 10.1186/s13046-023-02751-9 pmid: 37501099 |
| [23] | Li JY, Zhao Y, Gong S, Wang MM, Liu X, He QM, Li YQ, Huang SY, Qiao H, Tan XR, Ye ML, Zhu XH, He SW, Li Q, Liang YL, Chen KL, Huang SW, Li QJ, Ma J, Liu N. TRIM21 inhibits irradiation-induced mitochondrial DNA release and impairs antitumour immunity in nasopharyngeal carcinoma tumour models. Nat Commun, 2023, 14(1): 865. |
| [24] | Szántó M, Gupte R, Kraus WL, Pacher P, Bai P. PARPs in lipid metabolism and related diseases. Prog Lipid Res, 2021, 84: 101117. |
| [25] | Park SY, Kang KB, Thapa N, Kim SY, Lee SJ, Kim IS. Requirement of adaptor protein GULP during stabilin-2- mediated cell corpse engulfment. J Biol Chem, 2008, 283(16): 10593-10600. |
| [26] |
Osada Y, Sunatani T, Kim IS, Nakanishi Y, Shiratsuchi A. Signalling pathway involving GULP, MAPK and Rac1 for SR-BI-induced phagocytosis of apoptotic cells. J Biochem, 2009, 145(3): 387-394.
doi: 10.1093/jb/mvn176 pmid: 19122200 |
| [27] |
Merthan L, Haller A, Thal DR, von Einem B, von Arnim CAF. The role of PTB domain containing adaptor proteins on PICALM-mediated APP endocytosis and localization. Biochem J, 2019, 476(14): 2093-2109.
doi: 10.1042/BCJ20180840 pmid: 31300465 |
| [28] |
Vivien Chiu WY, Koon AC, Ki Ngo JC, Edwin Chan HY, Lau KF. GULP1/CED-6 ameliorates amyloid-β toxicity in a Drosophila model of Alzheimer's disease. Oncotarget, 2017, 8(59): 99274-99283.
doi: 10.18632/oncotarget.20062 pmid: 29245900 |
| [29] |
Datta S, Nam HS, Hayashi M, Maldonado L, Goldberg R, Brait M, Sidransky D, Illei P, Baras A, Vij N, Hoque MO. Expression of GULP1 in bronchial epithelium is associated with the progression of emphysema in chronic obstructive pulmonary disease. Respir Med, 2017, 124: 72-78.
doi: S0954-6111(17)30032-X pmid: 28284325 |
| [30] |
Gong JY, Gaitanos TN, Luu O, Huang YY, Gaitanos L, Lindner J, Winklbauer R, Klein R. Gulp1 controls Eph/ephrin trogocytosis and is important for cell rearrangements during development. J Cell Biol, 2019, 218(10): 3455-3471.
doi: 10.1083/jcb.201901032 pmid: 31409653 |
| [31] |
Song G, Suzuki OT, Santos CM, Lucas AT, Wiltshire T, Zamboni WC. Gulp1 is associated with the pharmacokinetics of PEGylated liposomal doxorubicin (PLD) in inbred mouse strains. Nanomedicine, 2016, 12(7): 2007-2017.
doi: S1549-9634(16)30066-1 pmid: 27288666 |
| [32] | Chau DD, Yu ZC, Chan WWR, Yuqi Z, Chang RCC, Ngo JCK, Chan HYE, Lau KF. The cellular adaptor GULP1 interacts with ATG14 to potentiate autophagy and APP processing. Cell Mol Life Sci, 2024, 81(1): 323. |
| [33] |
Faralli JA, Desikan H, Peotter J, Kanneganti N, Weinhaus B, Filla MS, Peters DM. Genomic/proteomic analyses of dexamethasone-treated human trabecular meshwork cells reveal a role for GULP1 and ABR in phagocytosis. Mol Vis, 2019, 25: 237-254.
pmid: 31516309 |
| [1] | Yueyang Wu, Xiaoyan Zhou, Yufeng Wu, Ju Huang. Effects of functional defects in the NMD pathway on rice phenotype and transcriptome [J]. Hereditas(Beijing), 2024, 46(7): 540-551. |
| [2] | Mingjie Sun, Jiali Lu, Yue Pang. Lamprey——an excellent model for iron metabolism [J]. Hereditas(Beijing), 2024, 46(5): 387-397. |
| [3] | Heng Wei, Tianpeng Liu, Jihong He, Kongjun Dong, Ruiyu Ren, Lei Zhang, Yawei Li, Ziyi Hao, Tianyu Yang. Genome-wide identification of GRF transcription factors and their expression profile in stem meristem of broomcorn millet (Panicum miliaceum L.) [J]. Hereditas(Beijing), 2024, 46(3): 242-255. |
| [4] | Yuxin Wan, Xinyu Zhu, Yu Zhao, Na Sun, Tiantongfei Jiang, Juan Xu. Computational dissection of the regulatory mechanisms of aberrant metabolism in remodeling the microenvironment of breast cancer [J]. Hereditas(Beijing), 2024, 46(10): 871-885. |
| [5] | Shuyu Mao, Changrui Zhao, Chang Liu. The nuclear receptor REV-ERBα integrates circadian clock and energy metabolism [J]. Hereditas(Beijing), 2023, 45(2): 99-114. |
| [6] | Fang Wang, Yuebo Zhang, Qian Jiang, Yulong Yin, Bi’e Tan, Jiashun Chen. Analysis of transcriptome differences between subcutaneous and intramuscular adipose tissue of Ningxiang pigs [J]. Hereditas(Beijing), 2023, 45(12): 1147-1157. |
| [7] | Yan Guo, Lele Yang, Huayu Qi. Transcriptome analysis of mouse male germline stem cells reveals characteristics of mature spermatogonial stem cells [J]. Hereditas(Beijing), 2022, 44(7): 591-608. |
| [8] | Mengxiao Wang, Margaret S. Ho. Recent advances on the role of glia in physiological behaviors: insights from Drosophila melanogaster [J]. Hereditas(Beijing), 2022, 44(4): 300-312. |
| [9] | Yizhun Zeng, Tao Zhang, Ying Xu. Rapid assessment of circadian behavior in mice [J]. Hereditas(Beijing), 2022, 44(4): 346-357. |
| [10] | Haoqiang Zhao, Xiaofei Wang, Shaopei Gao. Progress on the functional role of oleosin gene family in plants [J]. Hereditas(Beijing), 2022, 44(12): 1128-1140. |
| [11] | Shanshan Wang, Wanyi Zhao, Huixiao Wu, Meng Shu, Jiaxin Yuan, Li Fang, Chao Xu. Research on the variants of FGFR1 and CEP290 genes in idiopathic hypogonadotropin hypogonadism [J]. Hereditas(Beijing), 2022, 44(10): 937-949. |
| [12] | Zhiquan Tang, Li Shi, Jing Xiong. Progress of solute carrier SLC family in nonalcoholic fatty liver disease [J]. Hereditas(Beijing), 2022, 44(10): 881-898. |
| [13] | Enquan Zhang, Weicong Cai, Guiling Li, Jian Li, Jingwen Liu. Analysis of microRNA expression profile in Emiliania huxleyi in response to virus infection [J]. Hereditas(Beijing), 2021, 43(11): 1088-1100. |
| [14] | Yuanyuan Hao, Xiangqian Zhao, Fudeng Huang, Chunshou Li. The role of PPR proteins in posttranscriptional regulation of organelle components in plants [J]. Hereditas(Beijing), 2021, 43(11): 1050-1065. |
| [15] | Hongbo Luo, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of a DNA methylation-driven transcriptional signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2020, 42(8): 775-787. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||