Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (8): 775-787.doi: 10.16288/j.yczz.20-139
• Research Article • Previous Articles Next Articles
Prognostic and predictive value of a DNA methylation-driven transcriptional signature in hepatocellular carcinoma
Hongbo Luo1, Pengbo Cao2( ), Gangqiao Zhou1,2(
), Gangqiao Zhou1,2( )
)
- 1. Guizhou University School of Medicine, Guiyang 550025, China
2. State Key Lab of Proteomics, National Center for Protein Sciences (Beijing), Institute of Radiation Medicine, Academy of Military Medical Sciences, Academy of Military Sciences, Beijing 100850, China;
-
Received:2020-05-18Revised:2020-07-10Online:2020-08-20Published:2020-07-10 -
Contact:Cao Pengbo,Zhou Gangqiao E-mail:birchcpb@163.com;zhougq114@126.com -
Supported by:Supported by the National S&T Major Project Nos(2018ZX10732202);Supported by the National S&T Major Project Nos(2017ZX10203205)
Cite this article
Hongbo Luo, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of a DNA methylation-driven transcriptional signature in hepatocellular carcinoma[J]. Hereditas(Beijing), 2020, 42(8): 775-787.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
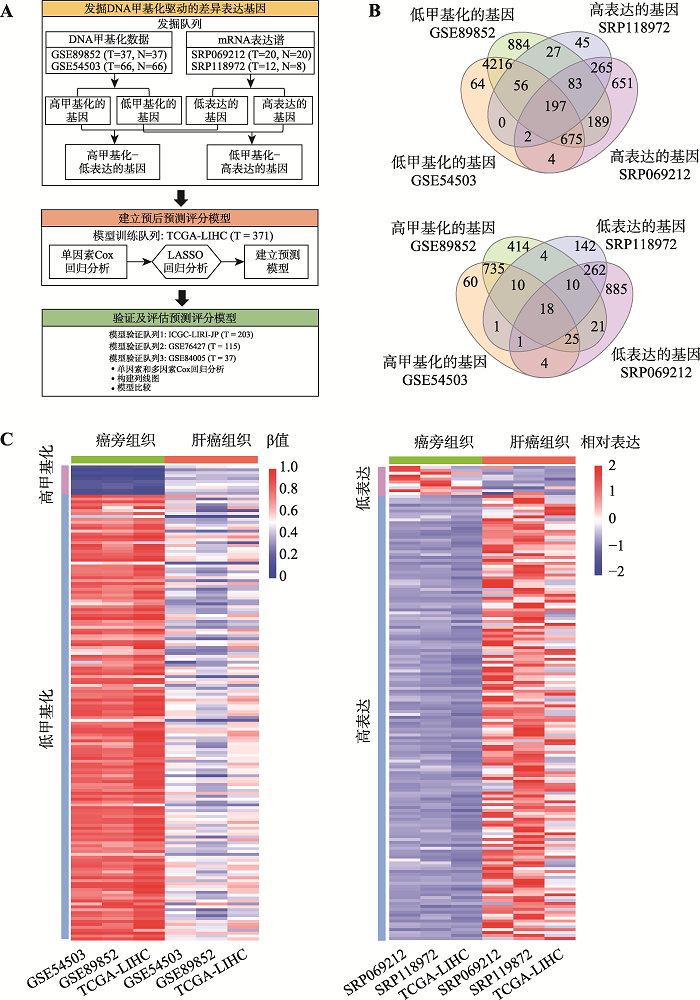
Samples and datasets used in this study"
| 研究队列 | 数据集 | 样本量 | 数据类型 | 数据来源 |
|---|---|---|---|---|
| 发掘队列 | SRP069212 | 20例配对的癌和癌旁组织 | mRNA表达 | GEO |
| SRP118972 | 12例癌组织样本和8例癌旁组织 | mRNA表达 | GEO | |
| GSE89852 | 33例配对的癌和癌旁组织 | DNA甲基化 | GEO | |
| GSE54503 | 66例配对的癌和癌旁组织 | DNA甲基化 | GEO | |
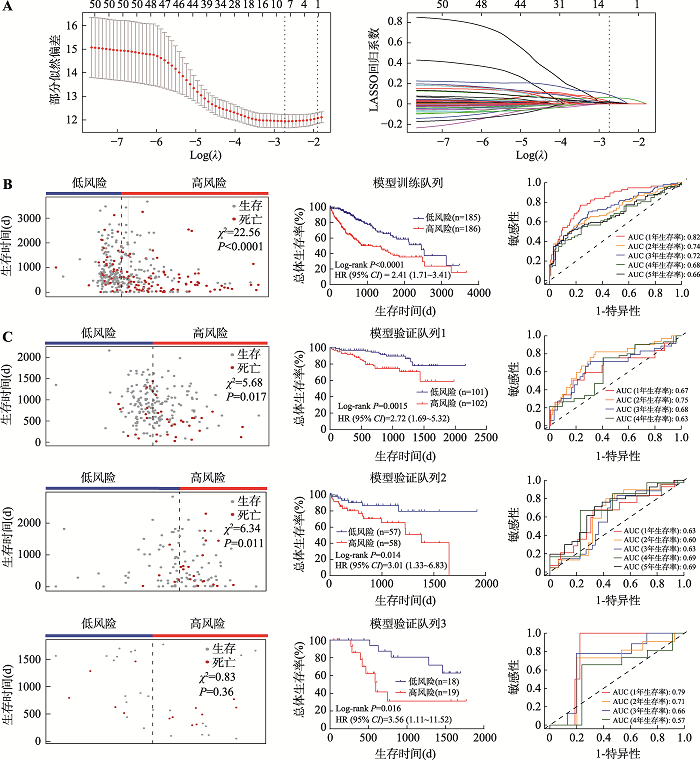
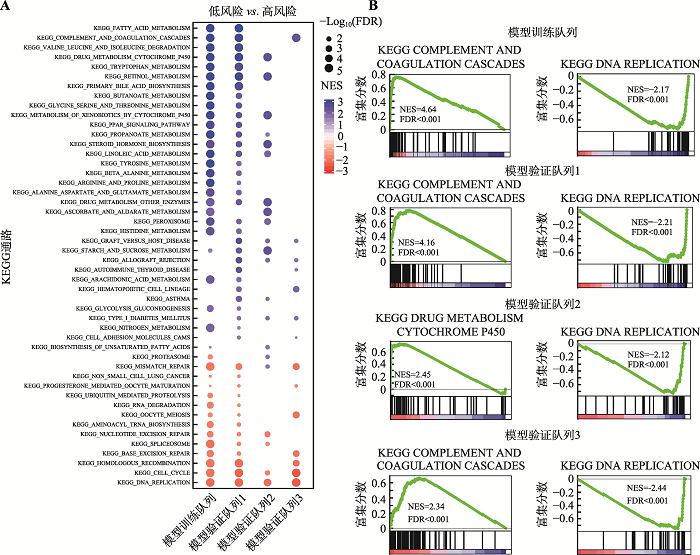
| 模型训练队列 | TCGA-LIHC | 371例癌组织和50例癌旁组织 | mRNA表达和DNA甲基化 | TCGA |
| ICGC-LIRI-JP | 203例癌组织 | mRNA表达 | ICGC | |
| 模型验证队列 | GSE76427 | 115例癌组织 | 基因表达 | GEO |
| GSE84005 | 37例癌组织 | 基因表达 | GEO |
Table 2
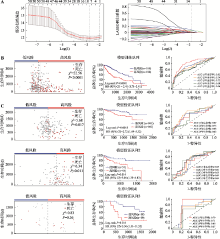
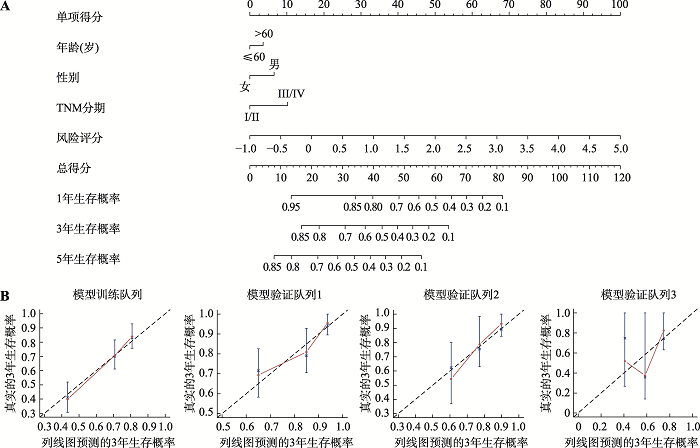
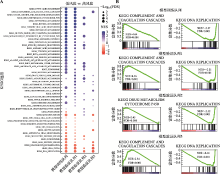
The results of Cox proportional hazard model analysis, LASSO regression coefficients and genetic alteration of the 10 optimal genes"
| 基因 | HR (95% CI) | P值 | LASSO系数 | 基因组变异频率(%) |
|---|---|---|---|---|
| CDCA8 | 2.21 (1.54~3.01) | <0.0001 | 0.1194 | 0.0 |
| PRC1 | 1.85 (1.29~2.54) | 0.0005 | 0.08869 | 0.3 |
| MAPT | 1.72 (1.21~2.46) | 0.0021 | 0.2597 | 1.7 |
| SFN | 1.72 (1.21~2.45) | 0.0021 | 0.001652 | 0.3 |
| STC2 | 1.73 (1.22~2.43) | 0.0021 | 0.03600 | 0.8 |
| MYO18B | 1.69 (1.19~2.37) | 0.0031 | 0.1932 | 4.0 |
| PBK | 1.67 (1.17~2.35) | 0.0036 | 0.06524 | 6.0 |
| MAEL | 1.55 (1.09~2.17) | 0.013 | -0.1766 | 10.0 |
| TTC39A | 1.55 (1.09~2.17) | 0.013 | -0.01347 | 1.4 |
| LPL | 1.55 (1.10~2.18) | 0.014 | 0.003126 | 7.0 |
| [1] |
Villanueva A . Hepatocellular carcinoma. N Engl J Med, 2019,380(15):1450-1462.
doi: 10.1056/NEJMra1713263 pmid: 30970190 |
| [2] |
Samonakis DN, Kouroumalis EA . Systemic treatment for hepatocellular carcinoma: still unmet expectations. World J Hepatol, 2017,9(2):80-90.
doi: 10.4254/wjh.v9.i2.80 pmid: 28144389 |
| [3] |
Zhang JW, Xu Q, Li GL . Epigenetics in the genesis and development of cancers. Hereditas(Beijing), 2019,41(7):567-581.
doi: 10.16288/j.yczz.19-077 pmid: 31307967 |
|
张競文, 续倩, 李国亮 . 癌症发生发展中的表观遗传学研究. 遗传, 2019,41(7):567-581.
doi: 10.16288/j.yczz.19-077 pmid: 31307967 |
|
| [4] |
Sun LY, Li XY, Sun ZW . Progress of epigenetics and its therapeutic application in hepatocellular carcinoma. Hereditas(Beijing), 2015,37(6):517-527.
doi: 10.16288/j.yczz.14-443 pmid: 26351047 |
|
孙凌云, 李星逾, 孙志为 . 原发性肝癌的表观遗传学及其治疗. 遗传, 2015,37(06):517-527.
doi: 10.16288/j.yczz.14-443 pmid: 26351047 |
|
| [5] |
Tan AC, Jimeno A, Lin SH, Wheelhouse J, Chan F, Solomon A, Rajeshkumar NV, Rubio-Viqueira B, Hidalgo M . Characterizing DNA methylation patterns in pancreatic cancer genome. Mol Oncol, 2009,3(5-6):425-438.
doi: 10.1016/j.molonc.2009.03.004 pmid: 19497796 |
| [6] |
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR . STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 2013,29(1):15-21.
doi: 10.1093/bioinformatics/bts635 |
| [7] |
Kim D, Langmead B, Salzberg SL . HISAT: a fast spliced aligner with low memory requirements. Nat Methods, 2015,12(4):357-360.
doi: 10.1038/nmeth.3317 pmid: 25751142 |
| [8] |
Love MI, Huber W, Anders S . Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014,15(12):550.
doi: 10.1186/s13059-014-0550-8 pmid: 25516281 |
| [9] |
Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S . ChAMP: 450k chip analysis methylation pipeline. Bioinformatics, 2014,30(3):428-430.
doi: 10.1093/bioinformatics/btt684 |
| [10] |
Ritchie ME, Phipson B, Wu D, Hu YF, Law CW, Shi W, Smyth GK . Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res, 2015,43(7):e47.
doi: 10.1093/nar/gkv007 pmid: 25605792 |
| [11] |
Mah WC, Thurnherr T, Chow PKH, Chung AYF, Ooi LLPJ, Toh HC, Teh BT, Saunthararajah Y, Lee CGL . Methylation profiles reveal distinct subgroup of hepatocellular carcinoma patients with poor prognosis. PLoS One, 2014,9(8):e104158.
doi: 10.1371/journal.pone.0104158 pmid: 25093504 |
| [12] | Friedman J, Hastie T, Tibshirani R . Regularization paths for generalized linear models via coordinate descent. J Stat Softw, 2010,33(1):1-22. |
| [13] |
Blanche P, Dartigues JF, Jacqmin-Gadda H . Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med, 2013,32(30):5381-5397.
doi: 10.1002/sim.5958 pmid: 24027076 |
| [14] |
Steyerberg EWS, Vergouwe Y . Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J, 2014,35(29):1925-1931.
doi: 10.1093/eurheartj/ehu207 |
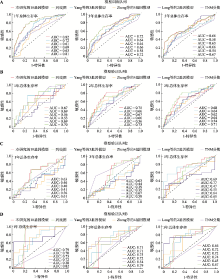
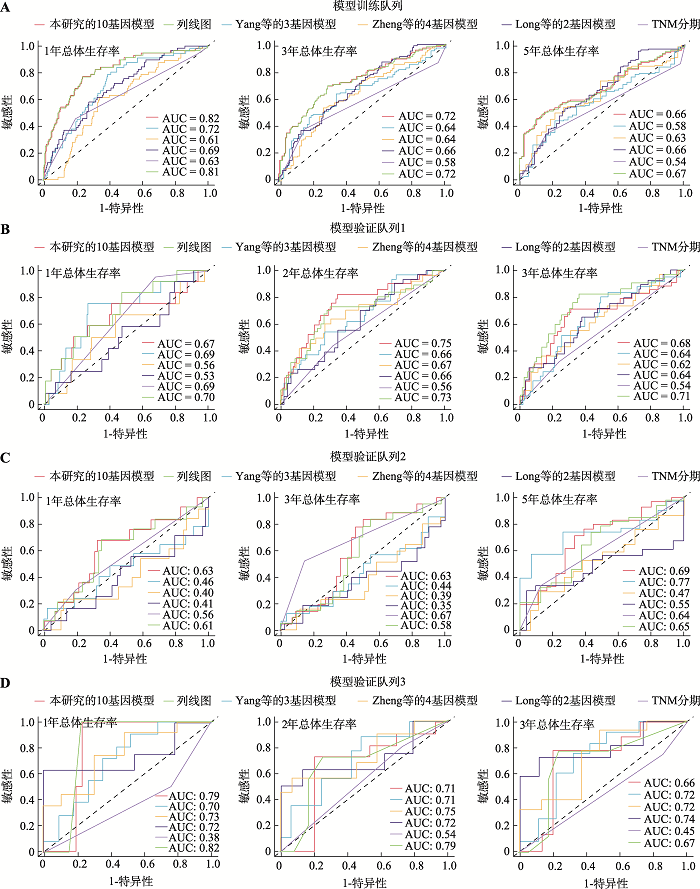
| [15] | Zheng YJ, Liu YL, Zhao SF, Zheng ZT, Shen CY, An L, Yuan YL . Large-scale analysis reveals a novel risk score to predict overall survival in hepatocellular carcinoma. Cancer Manag Res, 2018, ( 10):6079-6096. |
| [16] |
Yang Y, Lu Q, Shao XJ, Mo BH, Nie XQ, Liu W, Chen XH, Tang Y, Deng YC, Yan J . Development of a three-gene prognostic signature for hepatitis b virus associated heaptocellular carcinoma based on integrated transcriptomic analysis. J Cancer, 2018,9(11):1989-2002.
doi: 10.7150/jca.23762 pmid: 29896284 |
| [17] |
Long JY, Chen PP, Lin JZ, Bai Y, Yang X, Bian J, Lin Y, Wang DX, Yang XB, Zheng YC, Sang XT, Zhao HT . DNA methylation-driven genes for constructing diagnostic, prognostic, and recurrence models for hepatocellular carcinoma. Theranostics, 2019,9(24):7251-7267.
doi: 10.7150/thno.31155 pmid: 31695766 |
| [18] |
Torgovnick A, Schumacher B . DNA repair mechanisms in cancer development and therapy. Front Genet, 2015,6:157.
doi: 10.3389/fgene.2015.00157 pmid: 25954303 |
| [19] |
Feitelson MA . Parallel epigenetic and genetic changes in the pathogenesis of hepatitis virus-associated hepatocellular carcinoma. Cancer Lett, 2006,239(1):10-20.
doi: 10.1016/j.canlet.2005.07.009 pmid: 16154256 |
| [20] |
Liu LL, Dai YD, Chen JN, Zeng TT, Li Y, Chen LL, Zhu YH, Li JC, Li Y, Ma S, Xie D, Yuan YF, Guan XY . Maelstrom promotes hepatocellular carcinoma metastasis by inducing epithelial-mesenchymal transition by way of Akt/GSK-3β/Snail signaling. Hepatology, 2014,59(2):531-543.
doi: 10.1002/hep.26677 |
| [21] |
Zhang ZY, Zhu JF, Huang YS, Li WB, Cheng HQ . MYO18B promotes hepatocellular carcinoma progression by activating PI3K/AKT/mTOR signaling pathway. Diagn Pathol, 2018,13(1):85.
doi: 10.1186/s13000-018-0763-3 pmid: 30390677 |
| [22] |
Cao D, Song XH, Che L, Li XL, Pilo MG, Vidili G, Porcu A, Solinas A, Cigliano A, Pes GM, Ribback S, Dombrowski F, Chen X, Li L, Calvisi DF . Both de novo synthetized and exogenous fatty acids support the growth of hepatocellular carcinoma cells. Liver Int, 2017,37(1):80-89.
doi: 10.1111/liv.13183 pmid: 27264722 |
| [23] |
Liu P, Atkinson SJ, Akbareian SE, Zhou ZG, Munsterberg A, Robinson SD, Bao YP . Sulforaphane exerts anti-angiogenesis effects against hepatocellular carcinoma through inhibition of STAT3/HIF-1α/VEGF signalling. Sci Rep, 2017,7(1):12651.
doi: 10.1038/s41598-017-12855-w pmid: 28978924 |
| [24] | Wang HX, Wu KJ, Sun Y, Li YD, Wu MY, Qiao Q, Wei YJ, Han ZG, Cai B . STC2 is upregulated in hepatocellular carcinoma and promotes cell proliferation and migration in vitro. BMB Rep, 2012,45(11):629-634. |
| [25] |
Chen JX, Rajasekaran M, Xia HP, Zhang XQ, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BKP, Ooi LL, Hong WJ, Hui KM . The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut, 2016,65(9):1522-1534.
doi: 10.1136/gutjnl-2015-310625 pmid: 26941395 |
| [26] |
Desai A, Mitchison TJ . Microtubule polymerization dynamics. Annu Rev Cell Dev Biol, 1997,13:83-117.
doi: 10.1146/annurev.cellbio.13.1.83 pmid: 9442869 |
| [27] | Safran M, Dalah I, Alexander J, Rosen N, Stein TI, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, Sirota-Madi A, Olender T, Golan Y, Stelzer G, Harel A, Lancet D . GeneCards Version 3: the human gene integrator. Database (Oxford), 2010, 2010: baq020. |
| [1] | Mengxuan Xu, Ming Zhou. Advances of RNA polymerase IV in controlling DNA methylation and development in plants [J]. Hereditas(Beijing), 2022, 44(7): 567-580. |
| [2] | Yan Guo, Lele Yang, Huayu Qi. Transcriptome analysis of mouse male germline stem cells reveals characteristics of mature spermatogonial stem cells [J]. Hereditas(Beijing), 2022, 44(7): 591-608. |
| [3] | Min Cheng, Jing Zhang, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of the hypoxia-associated long non-coding RNA signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2022, 44(2): 153-167. |
| [4] | Changgui Lei, Xueyuan Jia, Wenjing Sun. Establish six-gene prognostic model for glioblastoma based on multi-omics data of TCGA database [J]. Hereditas(Beijing), 2021, 43(7): 665-679. |
| [5] | Tianpei Shi,Li Zhang. Application of whole transcriptomics in animal husbandry [J]. Hereditas(Beijing), 2019, 41(3): 193-205. |
| [6] | Gaohua Zhang, Shutao Yu, He Wang, Xuda Wang. Transcriptome profiling of high oleic peanut under low temperatureduring germination [J]. Hereditas(Beijing), 2019, 41(11): 1050-1059. |
| [7] | Yuwen Ke,Jiang Liu. The inheritance and reprogramming of chromatin structure in early animal embryos [J]. Hereditas(Beijing), 2018, 40(11): 977-987. |
| [8] | Lan Ren,Rudan Xiao,Qian Zhang,Xiaomin Lou,Zhaojun Zhang,Xiangdong Fang. Synergistic regulation of the erythroid differentiation of K562 cells by KLF1 and KLF9 [J]. Hereditas(Beijing), 2018, 40(11): 998-1006. |
| [9] | Yajun Liu,Feng Zhang,Hongde Liu,Xiao Sun. The application of next-generation sequencing techniques in studying transcriptional regulation in embryonic stem cells [J]. Hereditas(Beijing), 2017, 39(8): 717-725. |
| [10] | Kai Wei,Lei Ma. Concept development of housekeeping genes in the high-throughput sequencing era [J]. Hereditas(Beijing), 2017, 39(2): 127-134. |
| [11] | Guangqi Li, Congjiao Sun, Guiqin Wu, Fengying Shi, Aiqiao Liu, Hao Sun, Ning Yang. Transcriptome sequencing identifies potential regulatory genes involved in chicken eggshell brownness [J]. Hereditas(Beijing), 2017, 39(11): 1102-1111. |
| [12] | Yongming Liu, Ling Zhang, Tao Qiu, Zhuofan Zhao, Moju Cao. Research progress on mechanisms of male sterility in plants based on high-throughput RNA sequencing [J]. Hereditas(Beijing), 2016, 38(8): 677-687. |
| [13] | Xiao Zhang, Guifang Jia. RNA epigenetic modification: N6-methyladenosine [J]. HEREDITAS(Beijing), 2016, 38(4): 275-288. |
| [14] | Shuaiqi Zhu, Yifu Gong, Yuqing Hang, Hao Liu, Heyu Wang. Transcriptome analysis of Dunaliella viridis [J]. HEREDITAS(Beijing), 2015, 37(8): 828-836. |
| [15] | Dong Wang, Yongjun Li, Nan Ding, Junyun Wang, Qiong Yang, Yaran Yang, Yanming Li, Xiangdong Fang, Hua Zhao. Molecular networks and mechanisms of epithelial-mesenchymal transition regulated by miRNAs in the malignant melanoma cell line [J]. HEREDITAS(Beijing), 2015, 37(7): 673-682. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||