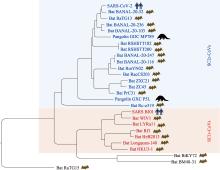
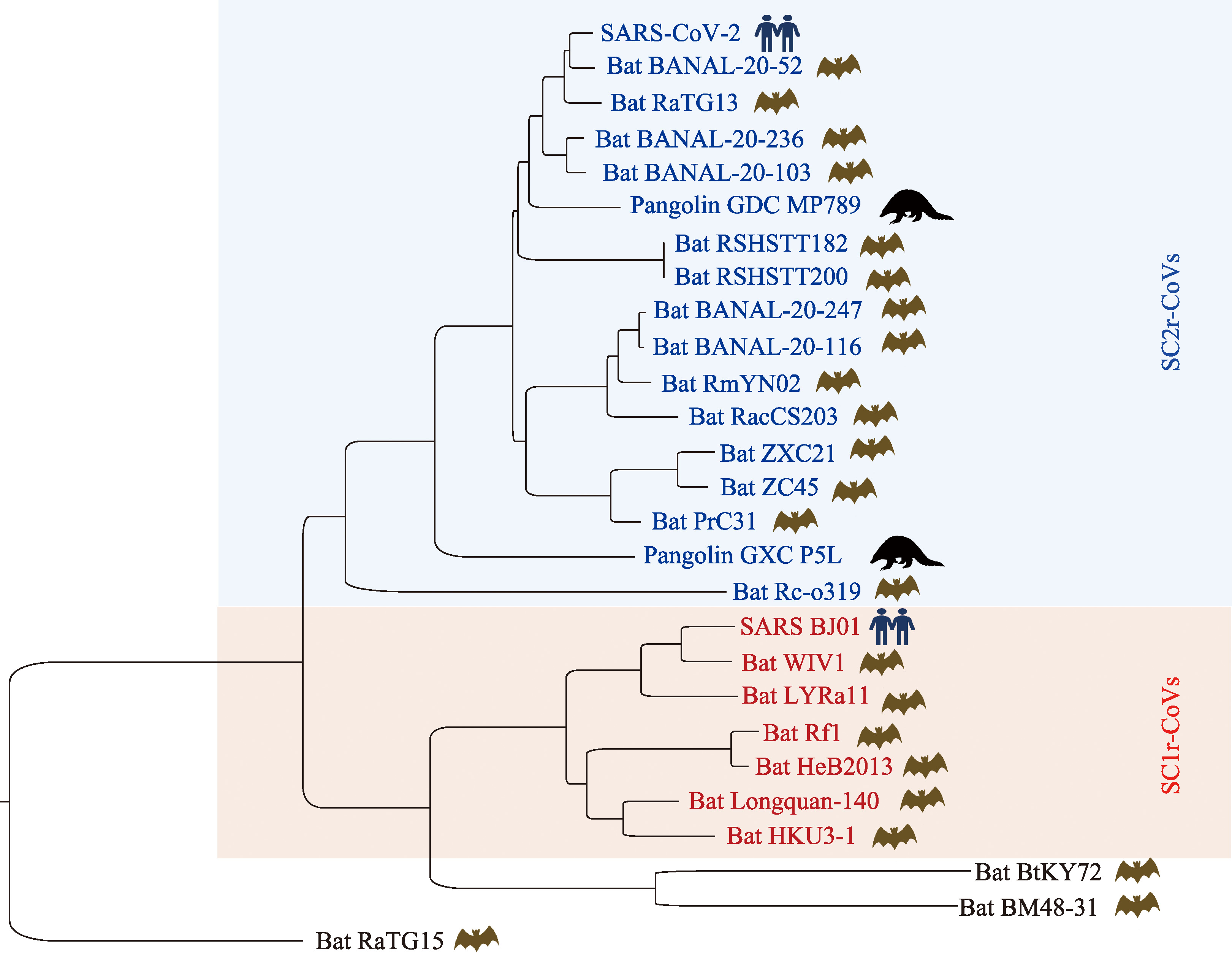
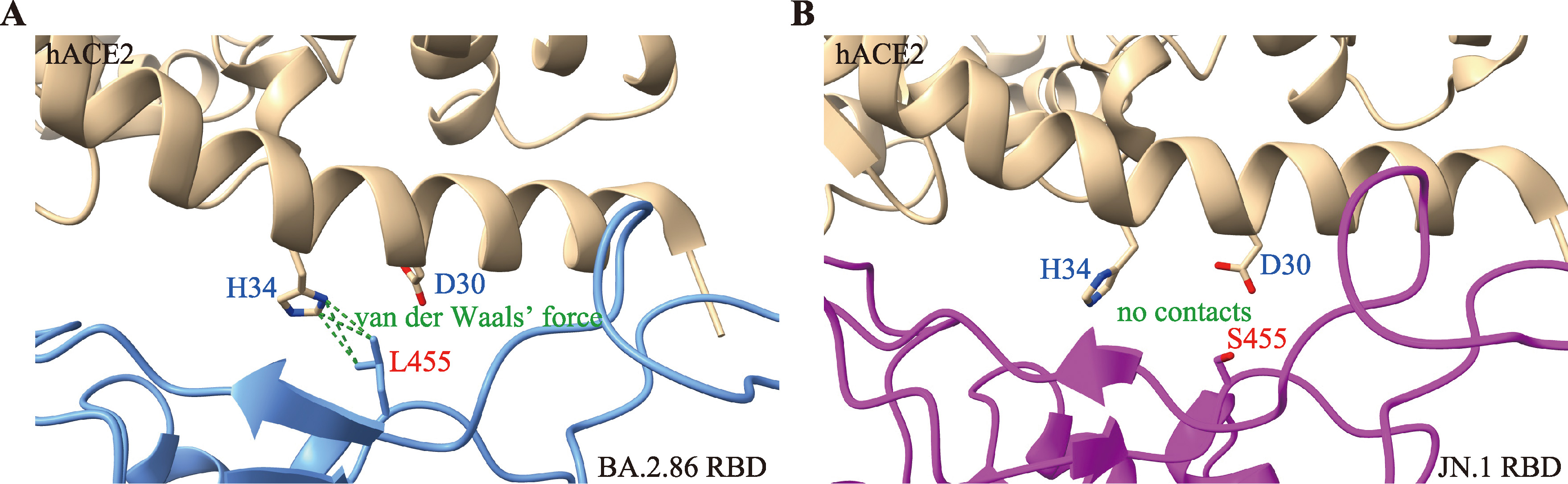
Hereditas(Beijing) ›› 2025, Vol. 47 ›› Issue (2): 211-227.doi: 10.16288/j.yczz.24-231
• Review • Previous Articles Next Articles
Current understanding of the adaptive evolution of the SARS-CoV-2 genome
Lin Zhang( ), Zhuocheng Yao(
), Zhuocheng Yao( ), Jian Lu(
), Jian Lu( ), Xiaolu Tang(
), Xiaolu Tang( )
)
- State Key Laboratory of Protein and Plant Gene Research, Center for Bioinformatics, School of Life Sciences, Peking University, Beijing 100871, China
-
Received:2024-08-08Revised:2024-09-26Online:2025-02-20Published:2024-11-18 -
Contact:Jian Lu, Xiaolu Tang E-mail:allan_z@stu.pku.edu.cn;yao15705723159@163.com;luj@pku.edu.cn;tangxiaolu@pku.edu.cn -
Supported by:National Key Research and Development Projects of the Ministry of Science and Technology of the People's Republic of China(2021YFC2301300);National Key Research and Development Projects of the Ministry of Science and Technology of the People's Republic of China(2023YFC3041500);National Key Research and Development Projects of the Ministry of Science and Technology of the People's Republic of China(2021YFC0863400)
Cite this article
Lin Zhang, Zhuocheng Yao, Jian Lu, Xiaolu Tang. Current understanding of the adaptive evolution of the SARS-CoV-2 genome[J]. Hereditas(Beijing), 2025, 47(2): 211-227.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Lu RJ, Zhao X, Li J, Niu PH, Yang B, Wu HL, Wang WL, Song H, Huang BY, Zhu N, Bi YH, Ma XJ, Zhan FX, Wang L, Hu T, Zhou H, Hu ZH, Zhou WM, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan JY, Xie ZH, Ma JM, Liu WJ, Wang DY, Xu WB, Holmes EC, Gao GF, Wu GZ, Chen WJ, Shi WF, Tan WJ. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet, 2020, 395(10224): 565-574.
doi: S0140-6736(20)30251-8 pmid: 32007145 |
| [2] | Ren LL, Wang YM, Wu ZQ, Xiang ZC, Guo L, Xu T, Jiang YZ, Xiong Y, Li YJ, Li XW, Li H, Fan GH, Gu XY, Xiao Y, Gao H, Xu JY, Yang F, Wang XM, Wu C, Chen L, Liu YW, Liu B, Yang J, Wang XR, Dong J, Li L, Huang CL, Zhao JP, Hu Y, Cheng ZS, Liu LL, Qian ZH, Qin C, Jin Q, Cao B, Wang JW. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl), 2020, 133(9): 1015-1024. |
| [3] | Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature, 2020, 579(7798): 265-269. |
| [4] | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 2020, 579(7798): 270-273. |
| [5] |
Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell, 2020, 181(4): 914-921.e10.
doi: S0092-8674(20)30406-2 pmid: 32330414 |
| [6] |
Parker MD, Lindsey BB, Leary S, Gaudieri S, Chopra A, Wyles M, Angyal A, Green LR, Parsons P, Tucker RM, Brown R, Groves D, Johnson K, Carrilero L, Heffer J, Partridge DG, Evans C, Raza M, Keeley AJ, Smith N, Filipe ADS, Shepherd JG, Davis C, Bennett S, Sreenu VB, Kohl A, Aranday-Cortes E, Tong L, Nichols J, Thomson EC, COVID-19 Genomics UK (COG-UK) Consortium, Wang D, Mallal S, de Silva TI. Subgenomic RNA identification in SARS-CoV-2 genomic sequencing data. Genome Res, 2021, 31(4): 645-658.
doi: 10.1101/gr.268110.120 pmid: 33722935 |
| [7] | Masters PS, Perlman S. Coronaviridae. In: Knipe DM, Howley PM, eds. Fields Virology. Sixth edn. 2013, 825-858. |
| [8] |
Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a Clinically Proven Protease Inhibitor. Cell, 2020, 181(2): 271-280.e8.
doi: S0092-8674(20)30229-4 pmid: 32142651 |
| [9] | Ou XY, Liu Y, Lei XB, Li P, Mi D, Ren LL, Guo L, Guo RX, Chen T, Hu JX, Xiang ZC, Mu ZX, Chen X, Chen JY, Hu KP, Jin Q, Wang JW, Qian ZH. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun, 2020, 11(1): 1620. |
| [10] |
Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med, 2020, 26(4): 450-452.
doi: 10.1038/s41591-020-0820-9 pmid: 32284615 |
| [11] | Wu CI, Wen HJ, Lu J, Su XD, Hughes AC, Zhai WW, Chen C, Chen H, Li MK, Song SH, Qian ZH, Wang QH, Chen BJ, Guo ZX, Ruan YS, Lu XM, Wei FW, Jin L, Kang L, Xue YB, Zhao GP, Zhang YP. On the origin of SARS-CoV-2--The blind watchmaker argument. Sci China Life Sci, 2021, 64(9): 1560-1563. |
| [12] |
Holmes EC, Goldstein SA, Rasmussen AL, Robertson DL, Crits-Christoph A, Wertheim JO, Anthony SJ, Barclay WS, Boni MF, Doherty PC, Farrar J, Geoghegan JL, Jiang XW, Leibowitz JL, Neil SJD, Skern T, Weiss SR, Worobey M, Andersen KG, Garry RF, Rambaut A. The origins of SARS-CoV-2: a critical review. Cell, 2021, 184(19): 4848-4856.
doi: 10.1016/j.cell.2021.08.017 pmid: 34480864 |
| [13] | Li J, Lai SJ, Gao GF, Shi WF. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature, 2021, 600(7889): 408-418. |
| [14] | Li WD, Shi ZL, Yu M, Ren WZ, Smith C, Epstein JH, Wang HZ, Crameri G, Hu ZH, Zhang HJ, Zhang JH, McEachern J, Field H, Daszak P, Eaton BT, Zhang SY, Wang LF. Bats are natural reservoirs of SARS- like coronaviruses. Science, 2005, 310(5748): 676-679. |
| [15] |
Dominguez SR, O'Shea TJ, Oko LM, Holmes KV. Detection of group 1 coronaviruses in bats in North America. Emerg Infect Dis, 2007, 13(9): 1295-1300.
doi: 10.3201/eid1309.070491 pmid: 18252098 |
| [16] |
Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol, 2019, 17(3): 181-192.
doi: 10.1038/s41579-018-0118-9 pmid: 30531947 |
| [17] | Temmam S, Vongphayloth K, Salazar EB, Munier S, Bonomi M, Régnault B, Douangboubpha B, Karami Y, Chretien D, Sanamxay D, Xayaphet V, Paphaphanh P, Lacoste V, Somlor S, Lakeomany K, Phommavanh N, Pérot P, Donati F, Bigot T, Nilges M, Rey F, van der Werf S, Brey P, Eloit M. Coronaviruses with a SARS-CoV-2-like receptor-binding domain allowing ACE2-mediated entry into human cells isolated from bats of Indochinese peninsula. Res Squire, 2021, doi:10.21203/rs.3.rs-871965/v1. |
| [18] | Zhou H, Chen X, Hu T, Li J, Song H, Liu YR, Wang PH, Liu D, Yang J, Holmes EC, Hughes AC, Bi YH, Shi WF. A novel bat coronavirus closely related to SARS- CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol, 2020, 30(11): 2196-2203.e3. |
| [19] |
Murakami S, Kitamura T, Suzuki J, Sato R, Aoi T, Fujii M, Matsugo H, Kamiki H, Ishida H, Takenaka-Uema A, Shimojima M, Horimoto T. Detection and characterization of bat Sarbecovirus phylogenetically related to SARS-CoV-2, Japan. Emerg Infect Dis, 2020, 26(12): 3025-3029.
doi: 10.3201/eid2612.203386 pmid: 33219796 |
| [20] |
Delaune D, Hul V, Karlsson EA, Hassanin A, Ou TP, Baidaliuk A, Gámbaro F, Prot M, Tu VT, Chea S, Keatts L, Mazet J, Johnson CK, Buchy P, Dussart P, Goldstein T, Simon-Lorière E, Duong V. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat Commun, 2021, 12(1): 6563.
doi: 10.1038/s41467-021-26809-4 pmid: 34753934 |
| [21] |
Zhou H, Ji JK, Chen X, Bi YH, Li J, Wang QH, Hu T, Song H, Zhao RC, Chen YH, Cui MX, Zhang YY, Hughes AC, Holmes EC, Shi WF. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell, 2021, 184(17): 4380-4391.e14.
doi: 10.1016/j.cell.2021.06.008 pmid: 34147139 |
| [22] |
Liu KF, Pan XQ, Li LJ, Yu F, Zheng AQ, Du P, Han PC, Meng YM, Zhang YF, Wu LL, Chen Q, Song CL, Jia YF, Niu S, Lu D, Qiao CP, Chen ZH, Ma DL, Ma XP, Tan SG, Zhao X, Qi JX, Gao GF, Wang QH. Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell, 2021, 184(13): 3438-3451.e10.
doi: 10.1016/j.cell.2021.05.031 pmid: 34139177 |
| [23] | Shang J, Ye G, Shi K, Wan YS, Luo CM, Aihara H, Geng QB, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature, 2020, 581(7807): 221-224. |
| [24] | Li P, Guo RX, Liu Y, Zhang YT, Hu JX, Ou XY, Mi D, Chen T, Mu ZX, Han YL, Chen ZH, Cui ZW, Zhang LL, Wang XQ, Wu ZQ, Wang JW, Jin Q, Qian ZH. The Rhinolophus affinis bat ACE2 and multiple animal orthologs are functional receptors for bat coronavirus RaTG13 and SARS-CoV-2. Sci Bull (Beijing), 2021, 66(12): 1215-1227. |
| [25] | Liu P, Jiang JZ, Wan XF, Hua Y, Li LM, Zhou JB, Wang XH, Hou FH, Chen J, Zou JJ, Chen JP. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog, 2020, 16(5): e1008421. |
| [26] |
Zhang T, Wu QF, Zhang ZG. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol, 2020, 30(7): 1346-1351.e2.
doi: S0960-9822(20)30360-2 pmid: 32197085 |
| [27] | Xiao KP, Zhai JQ, Feng YY, Zhou N, Zhang X, Zou JJ, Li N, Guo YQ, Li XB, Shen XJ, Zhang ZP, Shu FF, Huang WY, Li Y, Zhang ZD, Chen RA, Wu YJ, Peng SM, Huang M, Xie WJ, Cai QH, Hou FH, Chen W, Xiao LH, Shen YY. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature, 2020, 583(7815): 286-289. |
| [28] | Peng MS, Li JB, Cai ZF, Liu H, Tang XL, Ying RC, Zhang JN, Tao JJ, Yin TT, Zhang T, Hu JY, Wu RN, Zhou ZY, Zhang ZG, Yu L, Yao YG, Shi ZL, Lu XM, Lu J, Zhang YP. The high diversity of SARS-CoV-2- related coronaviruses in pangolins alerts potential ecological risks. Zool Res, 2021, 42(6): 834-844. |
| [29] | Lam TTY, Jia N, Zhang YW, Shum MHH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ, Jiang BG, Wei W, Yuan TT, Zheng K, Cui XM, Li J, Pei GQ, Qiang X, Cheung WYM, Li LF, Sun FF, Qin S, Huang JC, Leung GM, Holmes EC, Hu YL, Guan Y, Cao WC. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature, 2020, 583(7815): 282-285. |
| [30] | Wong MC, Javornik Cregeen SJ, Ajami NJ, Petrosino JF. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv, 2020, doi: 10.1101/2020.02.07.939207. |
| [31] |
Tang XL, Wu CC, Li X, Song YH, Yao XM, Wu XK, Duan YG, Zhang H, Wang YR, Qian ZH, Cui J, Lu J. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev, 2020, 7(6): 1012-1023.
doi: 10.1093/nsr/nwaa036 pmid: 34676127 |
| [32] | Niu S, Wang J, Bai B, Wu LL, Zheng AQ, Chen Q, Du P, Han PC, Zhang YF, Jia YF, Qiao CP, Qi JX, Tian WX, Wang HW, Wang QH, Gao GF. Molecular basis of cross-species ACE2 interactions with SARS-CoV- 2-like viruses of pangolin origin. EMBO J, 2021, 40(16): e107786. |
| [33] | Guo H, Hu B, Si HR, Zhu Y, Zhang W, Li B, Li A, Geng R, Lin HF, Yang XL, Zhou P, Shi ZL. Identification of a novel lineage bat SARS-related coronaviruses that use bat ACE2 receptor. Emerg Microbes Infect, 2021, 10(1): 1507-1514. |
| [34] | Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall, 2017, 1(1): 33-46. |
| [35] | Shu YL, McCauley J. GISAID: Global initiative on sharing all influenza data--from vision to reality. Euro Surveill, 2017, 22(13): 30494. |
| [36] | Yu WB, Tang GD, Zhang L, Corlett RT. Decoding the evolution and transmissions of the novel pneumonia coronavirus (SARS-CoV-2/HCoV-19) using whole genomic data. Zool Res, 2020, 41(3): 247-257. |
| [37] |
Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci USA, 2020, 117(17): 9241-9243.
doi: 10.1073/pnas.2004999117 pmid: 32269081 |
| [38] | Wu AP, Niu PH, Wang LL, Zhou HY, Zhao X, Wang WL, Wang JF, Ji CY, Ding X, Wang XY, Lu RJ, Gold S, Aliyari S, Zhang SL, Vikram E, Zou A, Lenh E, Chen J, Ye F, Han N, Peng YS, Guo HT, Wu GZ, Jiang TJ, Tan WJ, Cheng GH. Mutations, recombination and insertion in the evolution of 2019-nCoV. bioRxiv, 2020, doi: 10.1101/2020.02.29.971101. |
| [39] | Tang XL, Ying RC, Yao XM, Li GH, Wu CC, Tang YYL, Li ZD, Kuang BS, Wu F, Chi CS, Du XM, Qin Y, Gao SH, Hu SN, Ma JC, Liu TG, Pang XH, Wang JW, Zhao GP, Tan WJ, Zhang YP, Lu XM, Lu J. Evolutionary analysis and lineage designation of SARS-CoV-2 genomes. Sci Bull (Beijing), 2021, 66(22): 2297-2311. |
| [40] |
Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics, 2018, 34(23): 4121-4123.
doi: 10.1093/bioinformatics/bty407 pmid: 29790939 |
| [41] | Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, du Plessis L, Pybus OG. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol, 2020, 5(11): 1403-1407. |
| [42] | Cao YL, Wang J, Jian FC, Xiao TH, Song WL, Yisimayi A, Huang WJ, Li QQ, Wang P, An R, Wang J, Wang Y, Niu X, Yang SJ, Liang H, Sun HY, Li T, Yu YL, Cui QQ, Liu S, Yang XD, Du S, Zhang ZY, Hao XH, Shao F, Jin RH, Wang XX, Xiao JY, Wang YC, Xie XS. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature, 2022, 602(7898): 657-663. |
| [43] | Cao YL, Jian FC, Wang J, Yu YL, Song WL, Yisimayi A, Wang J, An R, Chen XS, Zhang N, Wang Y, Wang P, Zhao LJ, Sun HY, Yu LL, Yang SJ, Niu X, Xiao TH, Gu QQ, Shao F, Hao XH, Xu YL, Jin RH, Shen ZY, Wang YC, Xie XS. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature, 2023, 614(7948): 521-529. |
| [44] | Tang XL, Qian ZH, Lu XM, Lu J. Adaptive evolution of the Spike protein in coronaviruses. Mol Biol Evol, 2023, 40(4): msad089. |
| [45] |
Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, COVID-19 Genomics UK (COG-UK) Consortium, Peacock SJ, Robertson DL. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol, 2021, 19(7): 409-424.
doi: 10.1038/s41579-021-00573-0 pmid: 34075212 |
| [46] |
Carabelli AM, Peacock TP, Thorne LG, Harvey WT, Hughes J, COVID-19 Genomics UK Consortium, Peacock SJ, Barclay WS, de Silva TI, Towers GJ, Robertson DL. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol, 2023, 21(3): 162-177.
doi: 10.1038/s41579-022-00841-7 pmid: 36653446 |
| [47] |
Han PC, Li LJ, Liu S, Wang QS, Zhang D, Xu ZP, Han P, Li XM, Peng Q, Su C, Huang BH, Li DD, Zhang R, Tian MX, Fu LT, Gao YZ, Zhao X, Liu KF, Qi JX, Gao GF, Wang PY. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell, 2022, 185(4): 630-640.e10.
doi: 10.1016/j.cell.2022.01.001 pmid: 35093192 |
| [48] | Cantoni D, Murray MJ, Kalemera MD, Dicken SJ, Stejskal L, Brown G, Lytras S, Coey JD, McKenna J, Bridgett S, Simpson D, Fairley D, Thorne LG, Reuschl AK, Forrest C, Ganeshalingham M, Muir L, Palor M, Jarvis L, Willett B, Power UF, McCoy LE, Jolly C, Towers GJ, Doores KJ, Robertson DL, Shepherd AJ, Reeves MB, Bamford CGG, Grove J. Evolutionary remodelling of N-terminal domain loops fine-tunes SARS-CoV-2 spike. EMBO Rep, 2022, 23(10): e54322. |
| [49] |
Harbison AM, Fogarty CA, Phung TK, Satheesan A, Schulz BL, Fadda E. Fine-tuning the spike: role of the nature and topology of the glycan shield in the structure and dynamics of the SARS-CoV-2 S. Chem Sci, 2022, 13(2): 386-395.
doi: 10.1039/d1sc04832e pmid: 35126971 |
| [50] | Mittal A, Verma V. Connections between biomechanics and higher infectivity: a tale of the D614G mutation in the SARS-CoV-2 spike protein. Signal Transduct Target Ther, 2021, 6(1): 11. |
| [51] |
Liu YF, Soh WT, Kishikawa JI, Hirose M, Nakayama EE, Li SL, Sasai M, Suzuki T, Tada A, Arakawa A, Matsuoka S, Akamatsu K, Matsuda M, Ono C, Torii S, Kishida K, Jin H, Nakai W, Arase N, Nakagawa A, Matsumoto M, Nakazaki Y, Shindo Y, Kohyama M, Tomii K, Ohmura K, Ohshima S, Okamoto T, Yamamoto M, Nakagami H, Matsuura Y, Nakagawa A, Kato T, Okada M, Standley DM, Shioda T, Arase H. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell, 2021, 184(13): 3452-3466.e18.
doi: 10.1016/j.cell.2021.05.032 pmid: 34139176 |
| [52] | Wrobel AG, Benton DJ, Roustan C, Borg A, Hussain S, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. Evolution of the SARS-CoV-2 spike protein in the human host. Nat Commun, 2022, 13(1): 1178. |
| [53] | Liu Y, Liu JY, Johnson BA, Xia HJ, Ku ZQ, Schindewolf C, Widen SG, An ZQ, Weaver SC, Menachery VD, Xie XP, Shi PY. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep, 2022, 39(7): 110829. |
| [54] | Li C, Huang JJ, Yu YF, Wan ZX, Chiu MC, Liu XJ, Zhang SX, Cai JP, Chu H, Li G, Chan JFW, To KKW, Yang ZF, Jiang SB, Yuen KY, Clevers H, Zhou J. Human airway and nasal organoids reveal escalating replicative fitness of SARS-CoV-2 emerging variants. Proc Natl Acad Sci USA, 2023, 120(17): e2300376120. |
| [55] |
Baggen J, Jacquemyn M, Persoons L, Vanstreels E, Pye VE, Wrobel AG, Calvaresi V, Martin SR, Roustan C, Cronin NB, Reading E, Thibaut HJ, Vercruysse T, Maes P, De Smet F, Yee A, Nivitchanyong T, Roell M, Franco-Hernandez N, Rhinn H, Mamchak AA, Ah Young-Chapon M, Brown E, Cherepanov P, Daelemans D. TMEM106B is a receptor mediating ACE2- independent SARS-CoV-2 cell entry. Cell, 2023, 186(16): 3427-3442.e22.
doi: 10.1016/j.cell.2023.06.005 pmid: 37421949 |
| [56] | Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, Zhou S, Tuttle KS, Kim A, Li W, Dimitrov DS, Subramaniam S. Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding. Cell Rep, 2021, 37(12): 110156. |
| [57] |
Garcia-Beltran WF, Lam EC, St Denis K, Nitido AD, Garcia ZH, Hauser BM, Feldman J, Pavlovic MN, Gregory DJ, Poznansky MC, Sigal A, Schmidt AG, Iafrate AJ, Naranbhai V, Balazs AB. Multiple SARS-CoV-2 variants escape neutralization by vaccine- induced humoral immunity. Cell, 2021, 184(9): 2372-2383.e9.
doi: 10.1016/j.cell.2021.03.013 pmid: 33743213 |
| [58] |
Liu ZM, VanBlargan LA, Bloyet LM, Rothlauf PW, Chen RE, Stumpf S, Zhao HY, Errico JM, Theel ES, Liebeskind MJ, Alford B, Buchser WJ, Ellebedy AH, Fremont DH, Diamond MS, Whelan SPJ. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe, 2021, 29(3): 477-488.e4.
doi: 10.1016/j.chom.2021.01.014 pmid: 33535027 |
| [59] | Wang K, Jia ZJ, Bao LL, Wang L, Cao L, Chi H, Hu YL, Li QQ, Zhou YJ, Jiang YN, Zhu QH, Deng YQ, Liu P, Wang N, Wang L, Liu M, Li YR, Zhu BL, Fan KY, Fu WJ, Yang P, Pei XR, Cui Z, Qin LL, Ge PJ, Wu JJ, Liu S, Chen YD, Huang WJ, Wang Q, Qin CF, Wang YC, Qin C, Wang XX. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature, 2022, 603(7903): 919-925. |
| [60] |
Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, Wei Y, Atwal GS, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science, 2020, 369(6506): 1014-1018.
doi: 10.1126/science.abd0831 pmid: 32540904 |
| [61] | Weisblum Y, Schmidt F, Zhang FW, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann HH, Michailidis E, Gaebler C, Agudelo M, Cho A, Wang ZJ, Gazumyan A, Cipolla M, Luchsinger L, Hillyer CD, Caskey M, Robbiani DF, Rice CM, Nussenzweig MC, Hatziioannou T, Bieniasz PD. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife, 2020, 9: e61312. |
| [62] | Liu Y, Liu JY, Plante KS, Plante JA, Xie XP, Zhang XW, Ku ZQ, An ZQ, Scharton D, Schindewolf C, Widen SG, Menachery VD, Shi PY, Weaver SC. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature, 2022, 602(7896): 294-299. |
| [63] |
Gu HJ, Chen Q, Yang G, He L, Fan H, Deng YQ, Wang YX, Teng Y, Zhao ZP, Cui YJ, Li YC, Li XF, Li JF, Zhang NN, Yang XL, Chen SL, Guo Y, Zhao GY, Wang XL, Luo DY, Wang H, Yang X, Li Y, Han GC, He YX, Zhou XJ, Geng SS, Sheng XL, Jiang SB, Sun SH, Qin CF, Zhou YS. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science, 2020, 369(6511): 1603-1607.
doi: 10.1126/science.abc4730 pmid: 32732280 |
| [64] | Han PC, Su C, Zhang YF, Bai CZ, Zheng AQ, Qiao CP, Wang Q, Niu S, Chen Q, Zhang YQ, Li WW, Liao HY, Li J, Zhang ZY, Cho H, Yang MS, Rong XY, Hu Y, Huang N, Yan JH, Wang QH, Zhao X, Gao GF, Qi JX. Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants. Nat Commun, 2021, 12(1): 6103. |
| [65] | Zhu X, Mannar D, Srivastava SS, Berezuk AM, Demers JP, Saville JW, Leopold K, Li W, Dimitrov DS, Tuttle KS, Zhou S, Chittori S, Subramaniam S. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol, 2021, 19(4): e3001237. |
| [66] | Tian F, Tong B, Sun L, Shi SC, Zheng B, Wang ZB, Dong XC, Zheng P. N501Y mutation of spike protein in SARS-CoV-2 strengthens its binding to receptor ACE2. eLife, 2021, 10: e69091. |
| [67] | Sun SH, Gu HJ, Cao L, Chen Q, Ye Q, Yang G, Li RT, Fan H, Deng YQ, Song XP, Qi YN, Li M, Lan J, Feng R, Guo Y, Zhu N, Qin S, Wang L, Zhang YF, Zhou C, Zhao LN, Chen YH, Shen M, Cui YJ, Yang X, Wang XQ, Tan WJ, Wang H, Wang XX, Qin CF. Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2. Nat Commun, 2021, 12(1): 5654. |
| [68] | Kaku Y, Okumura K, Padilla-Blanco M, Kosugi Y, Uriu K, Hinay AA, Chen L, Plianchaisuk A, Kobiyama K, Ishii KJ, Genotype to Phenotype Japan (G2P-Japan) Consortium, Zahradnik J, Ito J, Sato K. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect Dis, 2024, 24(2): e82. |
| [69] | Yang SJ, Yu YL, Xu YL, Jian FC, Song WL, Yisimayi A, Wang P, Wang J, Liu JY, Yu LL, Niu X, Wang J, Wang Y, Shao F, Jin RH, Wang YC, Cao YL. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect Dis, 2024, 24(2): e70-e72. |
| [70] | Li LJ, Shi KY, Gu YH, Xu ZP, Shu C, Li DD, Sun JQ, Cong MQ, Li XM, Zhao X, Yu GH, Hu SN, Tan H, Qi JX, Ma XP, Liu KF, Gao GF. Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants. Structure, 2024, 32(8): 1055-1067.e6. |
| [71] | Focosi D, Maggi F. Neutralising antibody escape of SARS-CoV-2 spike protein: risk assessment for antibody-based Covid-19 therapeutics and vaccines. Rev Med Virol, 2021, 31(6): e2231. |
| [72] |
McCarthy KR, Rennick LJ, Nambulli S, Robinson- McCarthy LR, Bain WG, Haidar G, Duprex WP. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science, 2021, 371(6534): 1139-1142.
doi: 10.1126/science.abf6950 pmid: 33536258 |
| [73] |
McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu ZM, Zatta F, Zepeda S, di Iulio J, Bowen JE, Montiel-Ruiz M, Zhou JY, Rosen LE, Bianchi S, Guarino B, Fregni CS, Abdelnabi R, Foo SYC, Rothlauf PW, Bloyet LM, Benigni F, Cameroni E, Neyts J, Riva A, Snell G, Telenti A, Whelan SPJ, Virgin HW, Corti D, Pizzuto MS, Veesler D. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell, 2021, 184(9): 2332-2347.e16.
doi: 10.1016/j.cell.2021.03.028 pmid: 33761326 |
| [74] | Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, Wong KLM, van Zandvoort K, Silverman JD, CMMID COVID-19 Working Group, COVID-19 Genomics UK (COG-UK) ConsortiumU, Diaz-Ordaz K, Keogh R, Eggo RM, Funk S, Jit M, Atkins KE, Edmunds WJ. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science, 2021, 372(6538): eabg3055. |
| [75] |
Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O'Toole Á, Southgate J, Johnson R, Jackson B, Nascimento FF, Rey SM, Nicholls SM, Colquhoun RM, da Silva Filipe A, Shepherd J, Pascall DJ, Shah R, Jesudason N, Li K, Jarrett R, Pacchiarini N, Bull M, Geidelberg L, Siveroni I, COG-UK Consortium, Goodfellow I, Loman NJ, Pybus OG, Robertson DL, Thomson EC, Rambaut A, Connor TR. Evaluating the effects of SARS-CoV-2 Spike mutation D614G on transmissibility and pathogenicity. Cell, 2021, 184(1): 64-75.e11.
doi: 10.1016/j.cell.2020.11.020 pmid: 33275900 |
| [76] | Wang PF, Nair MS, Liu LH, Iketani S, Luo Y, Guo YC, Wang M, Yu J, Zhang BS, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng ZZ, Huang YX, Ho DD. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature, 2021, 593(7857): 130-135. |
| [77] | Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao NY, Korsman S, Davies MA, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Kosakovsky Pond SL, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature, 2021, 592(7854): 438-443. |
| [78] |
Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, Lambson BE, de Oliveira T, Vermeulen M, van der Berg K, Rossouw T, Boswell M, Ueckermann V, Meiring S, von Gottberg A, Cohen C, Morris L, Bhiman JN, Moore PL. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med, 2021, 27(4): 622-625.
doi: 10.1038/s41591-021-01285-x pmid: 33654292 |
| [79] |
Faria NR, Mellan TA, Whittaker C, Claro IM, da S Candido D, Mishra S, Crispim MAE, Sales FCS, Hawryluk I, McCrone JT, Hulswit RJG, Franco LAM, Ramundo MS, de Jesus JG, Andrade PS, Coletti TM, Ferreira GM, Silva CAM, Manuli ER, Pereira RHM, Peixoto PS, Kraemer MUG, Gaburo N, da C Camilo C, Hoeltgebaum H, Souza WM, Rocha EC, de Souza LM, de Pinho MC, Araujo LJT, Malta FSV, de Lima AB, do P Silva J, Zauli DAG, de S Ferreira AC, Schnekenberg RP, Laydon DJ, Walker PGT, Schlüter HM, Dos Santos ALP, Vidal MS, Del Caro VS, Filho RMF, Dos Santos HM, Aguiar RS, Proença-Modena JL, Nelson B, Hay JA, Monod M, Miscouridou X, Coupland H, Sonabend R, Vollmer M, Gandy A, Prete CA, Nascimento VH, Suchard MA, Bowden TA, Pond SLK, Wu CH, Ratmann O, Ferguson NM, Dye C, Loman NJ, Lemey P, Rambaut A, Fraiji NA, do P S S Carvalho M, Pybus OG, Flaxman S, Bhatt S, Sabino EC. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science, 2021, 372(6544): 815-821.
doi: 10.1126/science.abh2644 pmid: 33853970 |
| [80] |
Wang PF, Casner RG, Nair MS, Wang M, Yu J, Cerutti G, Liu LH, Kwong PD, Huang YX, Shapiro L, Ho DD. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe, 2021, 29(5): 747-751.e4.
doi: 10.1016/j.chom.2021.04.007 pmid: 33887205 |
| [81] | Zhang J, Xiao TS, Cai YF, Lavine CL, Peng HQ, Zhu HS, Anand K, Tong P, Gautam A, Mayer ML, Walsh RM, Rits-Volloch S, Wesemann DR, Yang W, Seaman MS, Lu JM, Chen B. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science, 2021, 374(6573): 1353-1360. |
| [82] | Li BS, Deng AP, Li KB, Hu Y, Li ZC, Xiong QL, Liu Z, Guo QF, Zou LR, Zhang H, Zhang M, Ouyang FZ, Su J, Su WZ, Xu J, Lin HF, Sun J, Peng JJ, Jiang HM, Zhou PP, Hu T, Luo M, Zhang YT, Zheng HY, Xiao JP, Liu T, Che RF, Zeng HR, Zheng ZH, Huang YS, Yu JX, Yi LN, Wu J, Chen JD, Zhong HJ, Deng XL, Kang M, Pybus OG, Hall M, Lythgoe KA, Li Y, Yuan J, He JF, Lu J. Viral infection and transmission in a large well-traced outbreak caused by the Delta SARS-CoV-2 variant. medRxiv, 2021, doi:10.1101/2021.07.07.21260122. |
| [83] | Zhang L, Li QQ, Liang ZT, Li T, Liu S, Cui QQ, Nie JH, Wu Q, Qu XW, Huang WJ, Wang YC. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg Microbes Infect, 2022, 11(1): 1-5. |
| [84] | Cele S, Jackson L, Khoury DS, Khan K, Moyo-Gwete T, Tegally H, San JE, Cromer D, Scheepers C, Amoako DG, Karim F, Bernstein M, Lustig G, Archary D, Smith M, Ganga Y, Jule Z, Reedoy K, Hwa SH, Giandhari J, Blackburn JM, Gosnell BI, Abdool Karim SS, Hanekom W, NGS-SA, COMMIT-KZN Team, von Gottberg A, Bhiman JN, Lessells RJ, Moosa MYS, Davenport MP, de Oliveira T, Moore PL, Sigal A. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature, 2022, 602(7898): 654-656. |
| [85] | Rössler A, Riepler L, Bante D, von Laer D, Kimpel J. SARS-CoV-2 Omicron variant neutralization in serum from vaccinated and convalescent persons. N Engl J Med, 2022, 386(7): 698-700. |
| [86] | Zhou YB, Zhi HL, Teng Y. The outbreak of SARS-CoV-2 Omicron lineages, immune escape, and vaccine effectivity. J Med Virol, 2023, 95(1): e28138. |
| [87] |
Willett BJ, Grove J, MacLean OA, Wilkie C, De Lorenzo G, Furnon W, Cantoni D, Scott S, Logan N, Ashraf S, Manali M, Szemiel A, Cowton V, Vink E, Harvey WT, Davis C, Asamaphan P, Smollett K, Tong L, Orton R, Hughes J, Holland P, Silva V, Pascall DJ, Puxty K, da Silva Filipe A, Yebra G, Shaaban S, Holden MTG, Pinto RM, Gunson R, Templeton K, Murcia PR, Patel AH, Klenerman P, Dunachie S, PITCH Consortium, COVID-19 Genomics UK (COG-UK) Consortium, Haughney J, Robertson DL, Palmarini M, Ray S, Thomson EC. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol, 2022, 7(8): 1161-1179.
doi: 10.1038/s41564-022-01143-7 pmid: 35798890 |
| [88] | Chen JH, Wang R, Gilby NB, Wei GW. Omicron (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance. ArXiv, 2021, doi: 10.48550/arXiv.2112.01318. |
| [89] | Wang Q, Guo YC, Iketani S, Nair MS, Li ZT, Mohri H, Wang M, Yu J, Bowen AD, Chang JY, Shah JG, Nguyen N, Chen ZW, Meyers K, Yin MT, Sobieszczyk ME, Sheng ZZ, Huang YX, Liu LH, Ho DD. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature, 2022, 608(7923): 603-608. |
| [90] | Liu LH, Iketani S, Guo YC, Chan JFW, Wang M, Liu LY, Luo Y, Chu H, Huang YM, Nair MS, Yu J, Chik KKH, Yuen TTT, Yoon C, To KKW, Chen HL, Yin MT, Sobieszczyk ME, Huang YX, Wang HH, Sheng ZZ, Yuen KY, Ho DD. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature, 2022, 602(7898): 676-681. |
| [91] | Verkhivker G, Agajanian S, Kassab R, Krishnan K. Computer simulations and network-based profiling of binding and allosteric interactions of SARS-CoV-2 Spike variant complexes and the host receptor: Dissecting the mechanistic effects of the Delta and Omicron mutations. Int J Mol Sci, 2022, 23(8): 4376. |
| [92] |
Singh A, Steinkellner G, Köchl K, Gruber K, Gruber CC. Serine 477 plays a crucial role in the interaction of the SARS-CoV-2 spike protein with the human receptor ACE2. Sci Rep, 2021, 11(1): 4320.
doi: 10.1038/s41598-021-83761-5 pmid: 33619331 |
| [93] | Yao ZC, Zhang L, Duan YG, Tang XL, Lu J. Molecular insights into the adaptive evolution of SARS-CoV-2 spike protein. J Infect, 2024, 88(3): 106121. |
| [94] |
Cui Z, Liu P, Wang N, Wang L, Fan KY, Zhu QH, Wang K, Chen RH, Feng R, Jia ZJ, Yang MN, Xu G, Zhu BL, Fu WJ, Chu TM, Feng LL, Wang YD, Pei XR, Yang P, Xie XS, Cao L, Cao YL, Wang XX. Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell, 2022, 185(5): 860-871.e13.
doi: 10.1016/j.cell.2022.01.019 pmid: 35120603 |
| [95] | Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, Kosugi Y, Shirakawa K, Sadamasu K, Kimura I, Ito J, Wu JQ, Iwatsuki-Horimoto K, Ito M, Yamayoshi S, Loeber S, Tsuda M, Wang L, Ozono S, Butlertanaka EP, Tanaka YL, Shimizu R, Shimizu K, Yoshimatsu K, Kawabata R, Sakaguchi T, Tokunaga K, Yoshida I, Asakura H, Nagashima M, Kazuma Y, Nomura R, Horisawa Y, Yoshimura K, Takaori-Kondo A, Imai M, Genotype to Phenotype Japan (G2P-Japan) Consortium, Tanaka S, Nakagawa S, Ikeda T, Fukuhara T, Kawaoka Y, Sato K. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature, 2022, 602(7896): 300-306. |
| [96] | Kozlov M. Omicron's feeble attack on the lungs could make it less dangerous. Nature, 2022, 601(7892): 177. |
| [97] | Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, Morioka Y, Nao N, Nasser H, Uriu K, Kosugi Y, Tsuda M, Orba Y, Sasaki M, Shimizu R, Kawabata R, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Genotype to Phenotype Japan (G2P-Japan) Consortium, Sawa H, Ikeda T, Irie T, Matsuno K, Tanaka S, Fukuhara T, Sato K. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature, 2022, 603(7902): 700-705. |
| [98] | Qu PK, Xu K, Faraone JN, Goodarzi N, Zheng YM, Carlin C, Bednash JS, Horowitz JC, Mallampalli RK, Saif LJ, Oltz EM, Jones D, Gumina RJ, Liu SL. Immune evasion, infectivity, and fusogenicity of SARS-CoV-2 BA.2.86 and FLip variants. Cell, 2024, 187(3): 585-595.e6. |
| [99] |
Zhang L, Kempf A, Nehlmeier I, Cossmann A, Richter A, Bdeir N, Graichen L, Moldenhauer AS, Dopfer-Jablonka A, Stankov MV, Simon-Loriere E, Schulz SR, Jäck HM, Čičin-Šain L, Behrens GMN, Drosten C, Hoffmann M, Pöhlmann S. SARS-CoV-2 BA.2.86 enters lung cells and evades neutralizing antibodies with high efficiency. Cell, 2024, 187(3): 596-608.e17.
doi: 10.1016/j.cell.2023.12.025 pmid: 38194966 |
| [100] | Teuwen LA, Geldhof V, Pasut A, Carmeliet P. COVID-19: the vasculature unleashed. Nat Rev Immunol, 2020, 20(7): 389-391. |
| [101] |
Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, Sheffield COVID-19 Genomics Group, McDanal C, Perez LG, Tang HL, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell, 2020, 182(4): 812-827.e19.
doi: S0092-8674(20)30820-5 pmid: 32697968 |
| [102] |
Chen NS, Zhou M, Dong X, Qu JM, Gong FY, Han Y, Qiu Y, Wang JL, Liu Y, Wei Y, Xia JA, Yu T, Zhang XX, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet, 2020, 395(10223): 507-513.
doi: S0140-6736(20)30211-7 pmid: 32007143 |
| [103] |
Wang DW, Hu B, Hu C, Zhu FF, Liu X, Zhang J, Wang BB, Xiang H, Cheng ZS, Xiong Y, Zhao Y, Li YR, Wang XH, Peng ZY. Clinical characteristics of 138 hospitalized patients with 2019 novel Coronavirus-infected pneumonia in Wuhan, China. JAMA, 2020, 323(11): 1061-1069.
doi: 10.1001/jama.2020.1585 pmid: 32031570 |
| [104] |
Stauft CB, Lien CZ, Selvaraj P, Liu SF, Wang TT. The G614 pandemic SARS-CoV-2 variant is not more pathogenic than the original D614 form in adult Syrian hamsters. Virology, 2021, 556: 96-100.
doi: 10.1016/j.virol.2021.01.005 pmid: 33556653 |
| [105] | Radvak P, Kwon HJ, Kosikova M, Ortega-Rodriguez U, Xiang RX, Phue JN, Shen RF, Rozzelle J, Kapoor N, Rabara T, Fairman J, Xie H. SARS-CoV-2 B.1.1.7 (alpha) and B.1.351 (beta) variants induce pathogenic patterns in K18-hACE2 transgenic mice distinct from early strains. Nat Commun, 2021, 12(1): 6559. |
| [106] | Chen Q, Huang XY, Tian Y, Fan CF, Sun MX, Zhou C, Li RT, Zhang RR, Wu GZ, Qin CF. The infection and pathogenicity of SARS-CoV-2 variant B.1.351 in hACE2 mice. Virol Sin, 2021, 36(5): 1232-1235. |
| [107] | Hu B, Liu R, Tang XL, Pan YC, Wang M, Tong YQ, Ye GM, Shen GG, Ying RC, Fu AS, Li D, Zhao WX, Peng J, Guo J, Men D, Yao XM, Wang YR, Zhang H, Feng ZH, Yu JP, Chen LJ, Deng ZX, Lu XM, Zhang YP, Li YR, Liu BD, Yu LL, Li Y, Lu J, Liu TG. The concordance between the evolutionary trend and the clinical manifestation of the two SARS-CoV-2 variants. Natl Sci Rev, 2021, 8(8): nwab073. |
| [108] | UK Health Security Agency. SARS-CoV-2 variants of concern and variants under investigation in England. 2021. https://assets.publishing.service.gov.uk/media/61c1c58bd3bf7f1f7ab31414/RA_Technical_Briefing_32_DRAFT_17_December_2021_2021_12_17.pdf. |
| [109] | Jalali N, Brustad HK, Frigessi A, MacDonald EA, Meijerink H, Feruglio SL, Nygård KM, Rø G, Madslien EH. Increased household transmission and immune escape of the SARS-CoV-2 Omicron compared to Delta variants. Nat Commun, 2022, 13(1): 5706. |
| [110] |
Lewnard JA, Hong VX, Patel MM, Kahn R, Lipsitch M, Tartof SY. Clinical outcomes associated with SARS- CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat Med, 2022, 28(9): 1933-1943.
doi: 10.1038/s41591-022-01887-z pmid: 35675841 |
| [111] | Abdullah F, Myers J, Basu D, Tintinger G, Ueckermann V, Mathebula M, Ramlall R, Spoor S, de Villiers T, Van der Walt Z, Cloete J, Soma-Pillay P, Rheeder P, Paruk F, Engelbrecht A, Lalloo V, Myburg M, Kistan J, van Hougenhouck-Tulleken W, Boswell MT, Gray G, Welch R, Blumberg L, Jassat W. Decreased severity of disease during the first global omicron variant covid-19 outbreak in a large hospital in tshwane, south africa. Int J Infect Dis, 2022, 116: 38-42. |
| [112] | Zhao HJ, Lu L, Peng Z, Chen LL, Meng XJ, Zhang CY, Ip JD, Chan WM, Chu AWH, Chan KH, Jin DY, Chen HL, Yuen KY, To KKW. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with delta variant in TMPRSS2- expressed cells. Emerg Microbes Infect, 2022, 11(1): 277-283. |
| [113] | Meng B, Ferreira IATM, Abdullahi A, Goonawardane N, Saito A, Kimura I, Yamasoba D, Gerba PP, Fatihi S, Rathore S, Zepeda SK, Papa G, Kemp SA, Ikeda T, Toyoda M, Tan TS, Kuramochi J, Mitsunaga S, Ueno T, Shirakawa K, Takaori-Kondo A, Brevini T, Mallery DL, Charles OJ, CITIID-NIHR BioResource COVID-19 Collaboration, The Genotype to Phenotype Japan (G2P-Japan) Consortium, Ecuador-COVID19 Consortium, Bowen JE, Joshi A, Walls AC, Jackson L, Cele S, Martin D, Smith KGC, Bradley J, Briggs JAG, Choi J, Madissoon E, Meyer K, Mlcochova P, Ceron-Gutierrez L, Doffinger R, Teichmann S, Pizzuto M, de Marco A, Corti D, Sigal A, James L, Veesler D, Hosmillo M, Lee JH, Sampaziotis F, Goodfellow IG, Matheson NJ, Thukral L, Sato K, Gupta RK. SARS-CoV-2 Omicron spike mediated immune escape and tropism shift. bioRxiv, 2022, doi: 10.1101/2021.12.17.473248. |
| [114] | Jin KC, Tang XL, Qian ZH, Wu ZQ, Yang ZF, Qian T, Hon C, Lu J. Modeling viral evolution: A novel SIRSVIDE framework with application to SARS-CoV-2 dynamics. hLife, 2024, 2(5): 227-245. |
| [115] |
Brian DA, Baric RS. Coronavirus genome structure and replication. Curr Top Microbiol Immunol, 2005, 287: 1-30.
pmid: 15609507 |
| [116] |
Lin XY, Sha Z, Trimpert J, Kunec D, Jiang C, Xiong Y, Xu BB, Zhu ZL, Xue WW, Wu HB. The NSP4 T492I mutation increases SARS-CoV-2 infectivity by altering non-structural protein cleavage. Cell Host Microbe, 2023, 31(7): 1170-1184.e7.
doi: 10.1016/j.chom.2023.06.002 pmid: 37402373 |
| [117] |
Sacco MD, Hu YM, Gongora MV, Meilleur F, Kemp MT, Zhang XJ, Wang J, Chen Y. The P132H mutation in the main protease of Omicron SARS-CoV-2 decreases thermal stability without compromising catalysis or small-molecule drug inhibition. Cell Res, 2022, 32(5): 498-500.
doi: 10.1038/s41422-022-00640-y pmid: 35292745 |
| [118] |
Goldswain H, Dong XF, Penrice-Randal R, Alruwaili M, Shawli GT, Prince T, Williamson MK, Raghwani J, Randle N, Jones B, Donovan-Banfield I, Salguero FJ, Tree JA, Hall Y, Hartley C, Erdmann M, Bazire J, Jearanaiwitayakul T, Semple MG, Openshaw PJM, Baillie JK, ISARIC4C Investigators, Emmett SR, Digard P, Matthews DA, Turtle L, Darby AC, Davidson AD, Carroll MW, Hiscox JA. The P323L substitution in the SARS-CoV-2 polymerase (NSP12) confers a selective advantage during infection. Genome Biol, 2023, 24(1): 47.
doi: 10.1186/s13059-023-02881-5 pmid: 36915185 |
| [119] | Arya R, Tripathi P, Nayak K, Ganesh J, Bihani SC, Ghosh B, Prashar V, Kumar M. Insights into the evolution of mutations in SARS-CoV-2 non-spike proteins. Microb Pathog, 2023, 185: 106460. |
| [120] |
Lu S, Ye QZ, Singh D, Cao Y, Diedrich JK, Yates JR, Villa E, Cleveland DW, Corbett KD. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat Commun, 2021, 12(1): 502.
doi: 10.1038/s41467-020-20768-y pmid: 33479198 |
| [121] | Syed AM, Taha TY, Tabata T, Chen IP, Ciling A, Khalid MM, Sreekumar B, Chen PY, Hayashi JM, Soczek KM, Ott M, Doudna JA. Rapid assessment of SARS-CoV- 2-evolved variants using virus-like particles. Science, 2021, 374(6575): 1626-1632. |
| [122] | Zhao HY, Nguyen A, Wu D, Li Y, Hassan SA, Chen JJ, Shroff H, Piszczek G, Schuck P. Plasticity in structure and assembly of SARS-CoV-2 nucleocapsid protein. PNAS Nexus, 2022, 1(2): pgac049. |
| [123] | Zhang JT, Ejikemeuwa A, Gerzanich V, Nasr M, Tang QY, Simard JM, Zhao RY. Understanding the role of SARS-CoV-2 ORF3a in viral pathogenesis and COVID-19. Front Microbiol, 2022, 13: 854567. |
| [124] | Addetia A, Lieberman NAP, Phung Q, Hsiang TY, Xie H, Roychoudhury P, Shrestha L, Loprieno MA, Huang ML, Gale M, Jerome KR, Greninger AL. SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98. mBio, 2021, 12(2): e00065-21. |
| [125] | Zhou ZL, Huang CL, Zhou ZC, Huang ZX, Su LL, Kang SS, Chen XX, Chen QY, He SH, Rong X, Xiao F, Chen J, Chen SD. Structural insight reveals SARS-CoV-2 ORF7a as an immunomodulating factor for human CD14+ monocytes. iScience, 2021, 24(3): 102187. |
| [126] | Li JY, Liao CH, Wang Q, Tan YJ, Luo R, Qiu Y, Ge XY. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res, 2020, 286: 198074. |
| [127] |
Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, Navarro MJ, Bowen JE, Tortorici MA, Walls AC, King NP, Veesler D, Bloom JD. Deep mutational scanning of SARS-CoV-2 receptor Binding domain reveals constraints on folding and ACE2 binding. Cell, 2020, 182(5): 1295-1310.e20.
doi: S0092-8674(20)31003-5 pmid: 32841599 |
| [128] |
Greaney AJ, Starr TN, Gilchuk P, Zost SJ, Binshtein E, Loes AN, Hilton SK, Huddleston J, Eguia R, Crawford KHD, Dingens AS, Nargi RS, Sutton RE, Suryadevara N, Rothlauf PW, Liu ZM, Whelan SPJ, Carnahan RH, Crowe JE, Bloom JD. Complete mapping of mutations to the SARS-CoV-2 Spike receptor-binding domain that escape antibody recognition. Cell Host Microbe, 2021, 29(1): 44-57.e9.
doi: 10.1016/j.chom.2020.11.007 pmid: 33259788 |
| [129] |
Dadonaite B, Crawford KHD, Radford CE, Farrell AG, Yu TC, Hannon WW, Zhou PP, Andrabi R, Burton DR, Liu LH, Ho DD, Chu HY, Neher RA, Bloom JD. A pseudovirus system enables deep mutational scanning of the full SARS-CoV-2 spike. Cell, 2023, 186(6): 1263-1278.e20.
doi: 10.1016/j.cell.2023.02.001 pmid: 36868218 |
| [130] | Wang GY, Liu XH, Wang K, Gao YX, Li G, Baptista-Hon DT, Yang XH, Xue KM, Tai WH, Jiang ZY, Cheng LL, Fok M, Lau JYN, Yang SY, Lu LG, Zhang P, Zhang K. Deep-learning-enabled protein-protein interaction analysis for prediction of SARS-CoV-2 infectivity and variant evolution. Nat Med, 2023, 29(8): 2007-2018. |
| [131] |
Han WK, Chen NN, Xu XZ, Sahil A, Zhou JX, Li ZX, Zhong HW, Gao E, Zhang RC, Wang Y, Sun SW, Cheung PPH, Gao X. Predicting the antigenic evolution of SARS-COV-2 with deep learning. Nat Commun, 2023, 14(1): 3478.
doi: 10.1038/s41467-023-39199-6 pmid: 37311849 |
| [132] |
Taft JM, Weber CR, Gao BC, Ehling RA, Han JM, Frei L, Metcalfe SW, Overath MD, Yermanos A, Kelton W, Reddy ST. Deep mutational learning predicts ACE2 binding and antibody escape to combinatorial mutations in the SARS-CoV-2 receptor-binding domain. Cell, 2022, 185(21): 4008-4022.e14.
doi: 10.1016/j.cell.2022.08.024 pmid: 36150393 |
| [1] | Zhongyi Sun, Guojie Zhang. Why should we stop translating “evolution” to “进化” and turn to use “演化” in Chinese [J]. Hereditas(Beijing), 2025, 47(1): 5-17. |
| [2] | Guilian Sheng, Mingmin Zheng, Bo Xiao, Junxia Yuan. Progress on ancient DNA investigation of Late Quaternary mammals in China [J]. Hereditas(Beijing), 2025, 47(1): 46-57. |
| [3] | Yuanfeng Li, Tianzhun Wu, Shunqi Chen, Yuting Wang, Tao Zeng, Ruofan Li, Gangqiao Zhou. Progresses on genetic susceptibility of COVID-19 [J]. Hereditas(Beijing), 2023, 45(11): 963-975. |
| [4] | Liqian Ren, Wei Liu, Wenbo Li, Wenjun Liu, Lei Sun. Peptidylprolyl cis/trans isomerase activity and molecular evolution of vertebrate Cyclophilin A [J]. Hereditas(Beijing), 2016, 38(8): 736-745. |
| [5] | Maomao Pu, Jun Yao, Xin Cao. Genomics: disclose the influence of human specific genetic variation on the evolution and development of cerebral cortex [J]. Hereditas(Beijing), 2016, 38(11): 957-970. |
| [6] | HUANG Yi-Min XIA Meng-Ying HUANG Shi. Evolutionary process unveiled by the maximum genetic diversity hypothesis [J]. HEREDITAS, 2013, 35(5): 599-606. |
| [7] | LIU Zhi-Xiang ZENG Chao-Zhen TAN Xiao-Feng. Molecular evolution of the poplar MIR169 gene family [J]. HEREDITAS, 2013, 35(11): 1307-1316. |
| [8] | WEI Jin-Pu, BO Hua-Feng, LI Gong-Quan, DUAN Fei. Distribution and evolution of simple repeats in the mtDNA D-loop in mammalian [J]. HEREDITAS, 2011, 33(1): 67-74. |
| [9] | NIE Qing-Hua, LIU Qing-Shen, FANG Mei-Xia, XIE Liang, ZHANG Xi-Quan. Analysis on molecular evolution of MC1R gene in dog [J]. HEREDITAS, 2008, 30(4): 469-474. |
| [10] | YU Hai-Long, XIAO Yun, AI Jing, LI Xia, GONG Bin-Sheng. Relationship between muscarinic acetylcholine receptor subtypes [J]. HEREDITAS, 2007, 29(10): 1280-1288. |
| [11] | SHAO Ai-Hua, ZHU Jiang, CHEN Kui, SHI Quan-Liang, YAO Wei-Wen. Charactreization and phylogenetic analysis of the CytochromOxidase Subunit I gene of Mitochondrial Genome from Takifugu fasciatus [J]. HEREDITAS, 2006, 28(8): 963-971. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||