Hereditas(Beijing) ›› 2025, Vol. 47 ›› Issue (7): 729-741.doi: 10.16288/j.yczz.24-374
• Review • Previous Articles Next Articles
Advances in microRNA promoting gene expression
Xiao Zhang( ), Yan Yu, Yong Ning, Qiwen Hong, Huaiping Shi(
), Yan Yu, Yong Ning, Qiwen Hong, Huaiping Shi( )
)
- College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China
-
Received:2024-12-30Revised:2025-02-28Online:2025-04-23Published:2025-04-23 -
Contact:Huaiping Shi E-mail:zhangzexiao@nwafu.edu.cn;huaipingshi@nwafu.edu.cn -
Supported by:National Key Research and Development Plan of China(2021YFD1600704);National Key Research and Development Plan of China(2022YFD1300200);National Natural Science Foundation of China(32272828)
Cite this article
Xiao Zhang, Yan Yu, Yong Ning, Qiwen Hong, Huaiping Shi. Advances in microRNA promoting gene expression[J]. Hereditas(Beijing), 2025, 47(7): 729-741.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
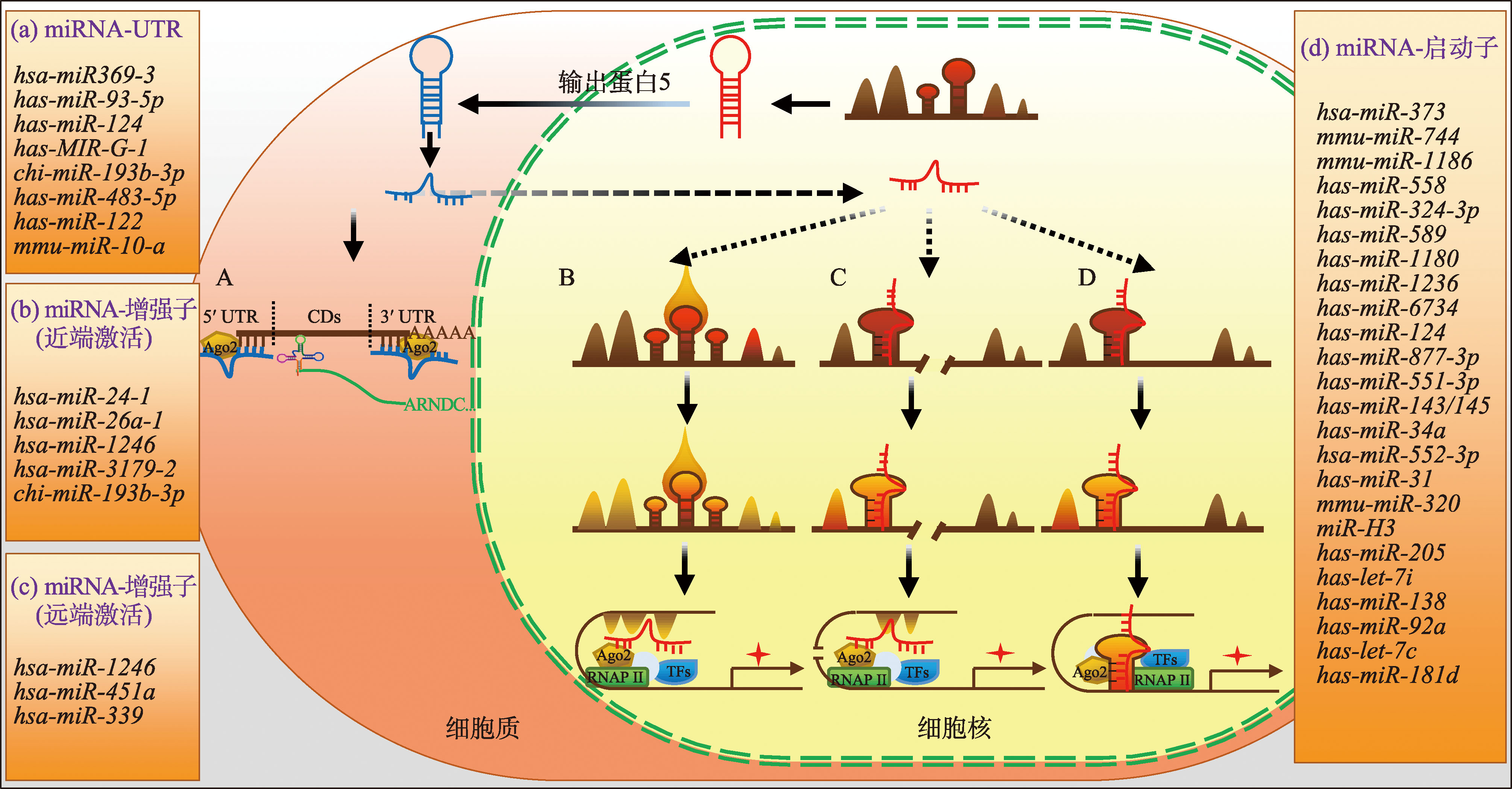
Table 1
Statistical results of miRNAs that promote gene expression via enhancer"
| 名称 | 靶基因 | “增强子-靶基因” 距离 | 参考 文献 |
|---|---|---|---|
| has-miR-24-1 | FBP1 | 482.88K | [ |
| FANCC | 13.03K | ||
| KDM6B | 283.33K | ||
| LSMD1 | 305.80K | ||
| CYB5D1 | 307.12K | ||
| has-miR-26a-1 | ITGA9 | 517.26K | [ |
| CTDSPL* | 107.52K | ||
| VILL | 18.57K | ||
| has-miR-1246 | MTX2 | 331.53K | [ |
| HNRNPA3 | 611.81K | ||
| HOXD3 | 448.45K | ||
| NFE2L2 | 629.32K | ||
| MMP1 | 70.00K | ||
| has-miR-3179-2 | ABCC6 | 150.59K | [ |
| PKD1P1 | 9.97K | ||
| chi-miR-193b-3p | MKL2 | 368.45K | [ |
| PARN | 112.35K | ||
| BFAR | 311.27K | ||
| hsa-miR-451a | KDM7A** | 28.22K | [ |
| has-miR-339 | GPER1 | 64.18K | [ |
Table 2
Statistical results of miRNAs that promote gene expression via promoter"
| 名称 | 靶向区域(相距转录起始位点) | 效应元件 | 生理效应 | 参考文献 |
|---|---|---|---|---|
| has-miR-373 | CSDC2(-787/-763) | 启动子 | ‒ | [ |
| CDH1(-645/-622) | 启动子 | |||
| mmu-miR-744 | Ccnb1(-192/-171) | 启动子 | 抑制细胞增殖,诱导染色质不稳定性 | [ |
| mmu-miR-1186 | Ccnb1(-699/-678) | 启动子 | ||
| has-miR-558 | HPSE(-2332/-2314) | 启动子 | 促进NB细胞生长、侵袭、转移和血管生成 | [ |
| has-miR-324-3p | RelA(-66/-45) | 启动子 | 诱导神经元细胞系凋亡 | [ |
| has-miR-589 | COX-2(-53/-37) | 启动子 | ‒ | [ |
| COX-2(-34/-14) | 启动子 | |||
| PLA2G4F(*/*) | 启动子 | |||
| has-miR-1180 | CDKN1A(-397/-379) | 启动子 | 抑制膀胱癌和肾癌细胞增殖,诱导细胞凋亡和衰老 | [ |
| has-miR-1236 | CDKN1A(-243/-226) | 启动子 | ||
| has-miR-6734 | CDKN1A(-322/-304) | 启动子 | 抑制结肠癌细胞系细胞增殖,诱导细胞周期阻滞 | [ |
| has-miR-124 | CDKN1B(-545/-533) | 启动子 | 抑制乳腺癌卵巢癌细胞增殖、迁移和侵袭,诱导细胞周期停滞 | [ |
| has-miR-877-3p | CDKN2A(-320/-299) | 启动子 | 抑制膀胱癌细胞增殖,诱导细胞周期停滞 | [ |
| has-miR-551-3p | STAT3(-530/-503) | 启动子 | [ | |
| has-miR-143/145 | SOX2(-1302/-1297) | 启动子 | 缓解雌激素缺乏型骨质流失 | [ |
| has-miR-34a | ZMYND10(-539/-341) (-98/-83)** | 启动子 | ‒ | [ |
| hsa-miR-552-3p | FXR(-1107/-1102) | 启动子 | 改善糖脂代谢紊乱 | [ |
| has-miR-31 | CD44(-890/-865) | 启动子 | 提高哮喘相关细胞因子 | [ |
| mmu-miR-320 | CD36(-553/-521) | 启动子 | 加剧糖尿病诱导的心功能障碍 | [ |
| miR-H3 | HIV-1 | 启动子(TATA盒) | 激活HIV病毒转录 | [ |
| has-miR-205 | IL24(-127/-107) | 启动子 | 促进前列腺癌细胞凋亡 | [ |
| IL32(-631/-610) | 启动子 | |||
| has-let-7i | IL2(201/222) | 启动子(TATA盒) | ‒ | [ |
| has-miR-138 | INS(-42/-18) | 启动子(TATA盒) | ||
| has-miR-92a | CALCA(-41/-17) | 启动子(TATA盒) | ||
| has-let-7c | H4A1(*/*) | 启动子(TATA盒) | ||
| has-miR-181d | MYC(-204/-175) | 启动子(TATA盒) |
Table 3
Statistical results of miRNAs that promote gene expression via UTR"
| 名称 | 靶向区域(相距起始密码子) | 效应元件 | 生理效应 | 参考文献 |
|---|---|---|---|---|
| has-miR369-3 | TNF-α(2,258/2,291) | 3′UTR (ARE) | 细胞周期停滞 | [ |
| has-miR-93-5p | MAP3K2(35,816/35,822) | 3′UTR | 促进HCC细胞增殖、迁移和侵袭 | [ |
| has-miR-124 | PTBP2(91,830/91,849)(92,569/92,595) | 3′UTR | 促进神经元功能成熟 | [ |
| has-MIR-G-1 | TMED5(27,557/27,581) | 3′UTR | 促进细胞增殖、迁移、侵袭和EMT进展 | [ |
| LMNB1(59,085/59,094) | 3′UTR | 促进DNA 损伤修复 | ||
| chi-miR-193b-3p | IGF2BP1(39,568/39,583) | 3′UTR | 促进肌肉发育 | [ |
| has-miR-483-5p | IGF2(-3,716/-3,214) | 5′UTR | 促进癌变 | [ |
| has-miR-122 | HCV-N(1B)(-329/-313) | 5′NCR | 促进丙肝病毒复制 | [ |
| mmu-miR-10-a | RPS16(-54/-26) | 5′UTR | 诱导致癌转化 | [ |
| [1] |
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell, 1993, 75(5): 843-854.
pmid: 8252621 |
| [2] |
Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell, 1993, 75(5): 855-862.
pmid: 8252622 |
| [3] |
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature, 2000, 403(6772): 901-906.
pmid: 10706289 |
| [4] |
Liang Y, Zou QP, Yu WQ. Steering against wind: a new network of NamiRNAs and enhancers. Genomics Proteomics Bioinformatics, 2017, 15(5): 331-337.
pmid: 28882787 |
| [5] |
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci USA, 2008, 105(5): 1608-1613.
pmid: 18227514 |
| [6] |
Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell, 2007, 128(6): 1105-1118.
pmid: 17382880 |
| [7] |
Ledford H. MicroRNAs won the Nobel--will they ever be useful as medicines? Nature, 2024, doi: 10.1038/d41586-024-03303-7.
pmid: 39384929 |
| [8] |
Raza A, Charagh S, Karikari B, Sharif R, Yadav V, Mubarik MS, Habib M, Zhuang YH, Zhang C, Chen H, Varshney RK, Zhuang WJ. miRNAs for crop improvement. Plant Physiol Biochem, 2023, 201: 107857.
pmid: 37437345 |
| [9] |
Wang X, Xu X, Ma Z, Huo YQ, Xiao ZT, Li Y, Wang YH. Dynamic mechanisms for pre-miRNA binding and export by exportin-5. RNA, 2011, 17(8): 1511-1528.
pmid: 21712399 |
| [10] | 杜秋丽. miRNA及其功能研究. 生物学通报, 2004, 39(8): 13-15. |
| [11] |
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 2005, 120(1): 15-20.
pmid: 15652477 |
| [12] |
Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res, 2009, 19(7): 1175-1183.
pmid: 19336450 |
| [13] |
Brümmer A, Hausser J. MicroRNA binding sites in the coding region of mRNAs: extending the repertoire of post-transcriptional gene regulation. Bioessays, 2014, 36(6): 617-626.
pmid: 24737341 |
| [14] | Liang Y. Dual roles of miRNAs: functions of activation and inhibition. Chinese Journal of Nature, 2017, 39(1): 31-36. |
| 梁英. miRNA: 除了抑制, 它还有激活功能. 自然杂志, 2017, 39(1): 31-36. | |
| [15] |
Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science, 2005, 309(5740): 1577-1581.
pmid: 16141076 |
| [16] |
Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell, 2008, 30(4): 460-471.
pmid: 18498749 |
| [17] |
Suzuki HI, Young RA, Sharp PA. Super-enhancer- mediated RNA processing revealed by integrative microRNA network analysis. Cell, 2017, 168(6): 1000-1014.
pmid: 28283057 |
| [18] |
Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science, 2007, 315(5808): 97-100.
pmid: 17204650 |
| [19] |
Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′trailers. PLoS One, 2010, 5(5): e10563.
pmid: 20498841 |
| [20] |
Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res, 2011, 39(13): 5682-5691.
pmid: 21427083 |
| [21] |
Xiao M, Li J, Li W, Wang Y, Wu FZ, Xi YP, Zhang L, Ding C, Luo HB, Li Y, Peng L, Zhao LP, Peng SL, Xiao Y, Dong SH, Cao J, Yu WQ. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol, 2017, 14(10): 1326-1334.
pmid: 26853707 |
| [22] |
Liang Y, Lu Q, Li W, Zhang DP, Zhang FL, Zou Q, Chen L, Tong Y, Liu MX, Wang SX, Li WX, Ren XG, Xu P, Yang ZC, Dong SH, Zhang BL, Huang YN, Li DQ, Wang HL, Yu WQ. Reactivation of tumour suppressor in breast cancer by enhancer switching through NamiRNA network. Nucleic Acids Res, 2021, 49(15): 8556-8572.
pmid: 34329471 |
| [23] |
Yang S, Zou QP, Liang Y, Zhang DP, Peng LN, Li W, Li WX, Liu MX, Tong Y, Chen L, Xu P, Yang ZC, Zhou KC, Xiao JR, Wang HL, Yu WQ. miR-1246 promotes osteosarcoma cell migration via NamiRNA-enhancer network dependent on Argonaute 2. MedComm (2020), 2024, 5(4): e543.
pmid: 38585233 |
| [24] |
Zhai PS, Tong T, Wang XN, Li CW, Liu C, Qin X, Li S, Xie F, Mao JY, Zhang JJ, Guo HY. Nuclear miR-451a activates KDM7A and leads to cetuximab resistance in head and neck squamous cell carcinoma. Cell Mol Life Sci, 2024, 81(1): 282.
pmid: 38943031 |
| [25] |
Li L, Zhang X, Yang HL, Xu XL, Chen Y, Dai DH, Zhan SY, Guo JZ, Zhong T, Wang LJ, Cao JX, Zhang HP. miR-193b-3p promotes proliferation of goat skeletal muscle satellite cells through activating IGF2BP1. Int J Mol Sci, 2022, 23(24): 15760.
pmid: 36555418 |
| [26] |
Li W, Yang S, Xu P, Zhang DP, Tong Y, Chen L, Jia B, Li A, Lian C, Ru DP, Zhang BL, Liu MX, Chen CC, Fu WH, Yuan SH, Gu CJ, Wang L, Li WX, Liang Y, Yang ZC, Ren XG, Wang SX, Zhang XY, Song YL, Xie YH, Lu HZ, Xu JQ, Wang HL, Yu WQ. SARS-CoV-2 RNA elements share human sequence identity and upregulate hyaluronan via NamiRNA-enhancer network. EBioMedicine, 2022, 76: 103861.
pmid: 35124429 |
| [27] |
Huang V, Place RF, Portnoy V, Wang J, Qi ZX, Jia ZJ, Yu A, Shuman M, Yu JW, Li LC. Upregulation of cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res, 2012, 40(4): 1695-1707.
pmid: 22053081 |
| [28] |
Qu HX, Zheng LD, Pu JR, Mei H, Xiang X, Zhao X, Li D, Li SW, Mao L, Huang K, Tong QS. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet, 2015, 24(9): 2539-2551.
pmid: 25616966 |
| [29] |
Dharap A, Pokrzywa C, Murali S, Pandi G, Vemuganti R. MicroRNA miR-324-3p induces promoter-mediated expression of RelA gene. PLoS One, 2013, 8(11): e79467.
pmid: 24265774 |
| [30] |
Matsui M, Chu YJ, Zhang HY, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res, 2013, 41(22): 10086-10109.
pmid: 23999091 |
| [31] |
Wang CH, Tang K, Li Z, Chen Z, Xu H, Ye ZQ. Targeted p21WAF1/CIP1 activation by miR-1236 inhibits cell proliferation and correlates with favorable survival in renal cell carcinoma. Urol Oncol, 2016, 34(2): 23-59.
pmid: 26421587 |
| [32] |
Kang MR, Park KH, Yang JO, Lee CW, Oh SJ, Yun J, Lee MY, Han SB, Kang JS. miR-6734 up-regulates p21 gene expression and induces cell cycle arrest and apoptosis in colon cancer cells. PLoS One, 2016, 11(8): e160961.
pmid: 27509128 |
| [33] |
Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T, Zhang F, Hill SM, Liu W, Maiti SN, Cooper L, Azencot R, Lopez-Berestein G, Rodriguez-Aguayo C, Roopaimoole R, Pecot CV, Sood AK, Mukherjee S, Gray JW, Mills GB, Ram PT. Functional proteomics identifies miRNAs to target a p27/Myc/phospho-Rb signature in breast and ovarian cancer. Oncogene, 2016, 35(6): 691-701.
pmid: 26865225 |
| [34] |
Li SQ, Zhu Y, Liang Z, Wang X, Meng S, Xu X, Xu XL, Wu J, Ji AL, Hu ZH, Lin YW, Chen H, Mao YQ, Wang W, Zheng XY, Liu B, Xie LP. Up-regulation of p16 by miR-877-3p inhibits proliferation of bladder cancer. Oncotarget, 2016, 7(32): 51773-51783.
pmid: 27429046 |
| [35] |
Chaluvally-Raghavan P, Jeong KJ, Pradeep S, Silva AM, Yu SX, Liu WB, Moss T, Rodriguez-Aguayo C, Zhang D, Ram P, Liu JS, Lu YL, Lopez-Berestein G, Calin GA, Sood AK, Mills GB. Direct upregulation of STAT3 by microRNA-551b-3p deregulates growth and metastasis of ovarian cancer. Cell Rep, 2016, 15(7): 1493-1504.
pmid: 27160903 |
| [36] |
Xu RY, Shen X, Xie HY, Zhang HG, Liu DS, Chen X, Fu Y, Zhang P, Yang Y, Cheng J, Jiang HB. Identification of the canonical and noncanonical role of miR-143/145 in estrogen-deficient bone loss. Theranostics, 2021, 11(11): 5491-5510.
pmid: 33859759 |
| [37] |
Ohno SI, Oikawa K, Tsurui T, Harada Y, Ono K, Tateishi M, Mirza A, Takanashi M, Kanekura K, Nagase K, Shimada Y, Kudo Y, Ikeda N, Ochiya T, Wang XZ, Kuroda M. Nuclear microRNAs release paused Pol II via the DDX21-CDK9 complex. Cell Rep, 2022, 39(2): 110673.
pmid: 35417682 |
| [38] |
Fan L, Lai RT, Ma NN, Dong YX, Li Y, Wu Q, Qiao JW, Lu HL, Gong LK, Tao ZT, Chen J, Xie Q, Ren J. miR-552-3p modulates transcriptional activities of FXR and LXR to ameliorate hepatic glycolipid metabolism disorder. J Hepatol, 2020, 74(1): 8-19.
pmid: 32818571 |
| [39] | Li L, Hui Y, Xing CF, Guo Y, Wang Q, Shu J, Qian J, Zhou GP. MicroRNA-31 affects the expression of asthma-related cytokines via regulation of CD44. Int J Clin Exp Med, 2016, 9(11): 21506-21513. |
| [40] |
Li HP, Fan JH, Zhao YR, Zhang XR, Dai BB, Zhan JB, Yin ZW, Nie X, Fu XD, Chen C, Wang DW. Nuclear miR-320 mediates diabetes-induced cardiac dysfunction by activating transcription of fatty acid metabolic genes to cause lipotoxicity in the heart. Circ Res, 2019, 125(12): 1106-1120.
pmid: 31638474 |
| [41] |
Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, Deng G, Dahiya R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer, 2010, 116(24): 5637-5649.
pmid: 20737563 |
| [42] |
Victoria P, Vera H, F PR, Long-Cheng L. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA, 2011, 2(5): 748-760.
pmid: 21823233 |
| [43] |
Kim DH, Saetrom P, Snøve O Jr, Rossi JJ. MicroRNA- directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA, 2008, 105(42): 16230-16235.
pmid: 18852463 |
| [44] |
Zhang YJ, Fan MM, Geng GN, Liu BF, Huang ZQ, Luo HH, Zhou J, Guo XM, Cai WP, Zhang H. A novel HIV-1-encoded microRNA enhances its viral replication by targeting the TATA box region. Retrovirology, 2014, 11(1): 23.
pmid: 24620741 |
| [45] |
Zhang YJ, Fan MM, Zhang X, Huang F, Wu K, Zhang JS, Liu J, Huang ZQ, Luo HH, Tao L, Zhang H. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA, 2014, 20(12): 1878-1889.
pmid: 25336585 |
| [46] |
Laitinen P, Väänänen MA, Kolari IL, Mäkinen PI, Kaikkonen MU, Weinberg MS, Morris KV, Korhonen P, Malm T, Ylä-Herttuala S, Roberts TC, Turunen MP, Turunen TA. Nuclear microRNA-466c regulates Vegfa expression in response to hypoxia. PLoS One, 2022, 17(3): e265948.
pmid: 19578358 |
| [47] |
Dikstein R. The unexpected traits associated with core promoter elements. Transcription, 2011, 2(5): 201-206.
pmid: 22231114 |
| [48] |
Vasudevan S, Tong YC, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science, 2007, 318(5858): 1931-1934.
pmid: 18048652 |
| [49] |
Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature, 2009, 460(7256): 705-710.
pmid: 19578358 |
| [50] |
Li HP, Zhang XR, Wang F, Zhou L, Yin ZW, Fan JH, Nie X, Wang PH, Fu XD, Chen C, Wang DW. MicroRNA-21 lowers blood pressure in spontaneous hypertensive rats by upregulating mitochondrial translation. Circulation, 2016, 134(10): 734-751.
pmid: 27542393 |
| [51] |
Shi X, Liu TT, Yu XN, Balakrishnan A, Zhu HR, Guo HY, Zhang GC, Bilegsaikhan E, Sun JL, Song GQ, Weng SQ, Dong L, Ott M, Zhu JM, Shen XZ. microRNA-93-5p promotes hepatocellular carcinoma progression via a microRNA-93-5p/MAP3K2/c-Jun positive feedback circuit. Oncogene, 2020, 39(35): 5768-5781.
pmid: 32719439 |
| [52] |
Reyes-Gutierrez P, Politz JCR, Pederson T. A mRNA and cognate microRNAs localize in the nucleolus. Nucleus, 2014, 5(6): 636-642.
pmid: 25485975 |
| [53] |
Lu YL, Liu YJ, Mccoy MJ, Yoo AS. MiR-124 synergism with ELAVL3 enhances target gene expression to promote neuronal maturity. Proc Natl Acad Sci USA, 2021, 118(22): 1-12.
pmid: 34031238 |
| [54] |
Yang Z, Sun Q, Guo JF, Wang SX, Song G, Liu WY, Liu M, Tang H. GRSF1-mediated MIR-G-1 promotes malignant behavior and nuclear autophagy by directly upregulating TMED5 and LMNB1 in cervical cancer cells. Autophagy, 2019, 15(4): 668-685.
pmid: 30394198 |
| [55] |
Liu MZ, Roth A, Yu M, Morris R, Bersani F, Rivera MN, Lu J, Shioda T, Vasudevan S, Ramaswamy S, Maheswaran S, Diederichs S, Haber DA. The IGF2 intronic miR-483 selectively enhances transcription from IGF2 fetal promoters and enhances tumorigenesis. Genes Dev, 2013, 27(23): 2543-2548.
pmid: 24298054 |
| [56] |
Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J, Trimbuch T, Eom G, Hinz M, Kaul D, Habbel P, Kälin R, Franzoni E, Rybak A, Nguyen D, Veh R, Ninnemann O, Peters O, Nitsch R, Heppner FL, Golenbock D, Schott E, Ploegh HL, Wulczyn FG, Lehnardt S. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci, 2012, 15(6): 827-835.
pmid: 22610069 |
| [57] |
Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA, 2012, 109(31): E2110-E2116.
pmid: 22753494 |
| [58] |
Lu WT, Bushell M. Old case, new leads: miRNA links Kaposi's sarcoma-associated herpesvirus with sepsis. Cell Death Dis, 2014, 5(12): e1560.
pmid: 25476908 |
| [59] |
Tudor S, Giza DE, Lin HY, Fabris L, Yoshiaki K, D'Abundo L, Toale KM, Shimizu M, Ferracin M, Challagundla KB, Cortez MA, Fuentes-Mattei E, Tulbure D, Gonzalez C, Henderson J, Row M, Rice TW, Ivan C, Negrini M, Fabbri M, Morris JS, Yeung SCJ, Vasilescu C, Calin GA. Cellular and Kaposi's sarcoma-associated herpes virus microRNAs in sepsis and surgical trauma. Cell Death Dis, 2014, 5(12): e1559.
pmid: 25476907 |
| [60] |
Zhang XR, Zuo XX, Yang B, Li ZR, Xue YC, Zhou Y, Huang J, Zhao XL, Zhou J, Yan Y, Zhang HQ, Guo PP, Sun H, Guo L, Zhang Y, Fu XD. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell, 2014, 158(3): 607-619.
pmid: 25083871 |
| [61] |
Matsui M, Sakurai F, Elbashir S, Foster DJ, Manoharan M, Corey DR. Activation of LDL receptor expression by small RNAs complementary to a noncoding transcript that overlaps the LDLR promoter. Chem Biol, 2010, 17(12): 1344-1355.
pmid: 21168770 |
| [62] |
Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, Tissières V, Pickle CS, Plajzer-Frick I, Lee EA, Kato M, Garvin TH, Akiyama JA, Afzal V, Lopez-Rios J, Rubin EM, Dickel DE, Pennacchio LA, Visel A. Progressive loss of function in a limb enhancer during snake evolution. Cell, 2016, 167(3): 633-642.
pmid: 27768887 |
| [63] | Cheng X, Yang Q, Tan ZD, Tan Y, Pu HZ, Zhao X, Zhang SH, Zhu L. The current research status of enhancer RNAs. Hereditas(Beijing), 2017, 39(9): 784-797. |
| 程霄, 杨琼, 谭镇东, 谭娅, 蒲红州, 赵雪, 张顺华, 朱砺. 增强子RNA研究现状. 遗传, 2017, 39(9): 784-797. | |
| [64] |
Bose DA, Donahue G, Reinberg D, Shiekhattar R, Bonasio R, Berger SL. RNA binding to CBP stimulates histone acetylation and transcription. Cell, 2017, 168(1-2): 135-149.
pmid: 28086087 |
| [65] |
Nishi K, Nishi A, Nagasawa T, Ui-Tei K. Human TNRC6A is an Argonaute-navigator protein for microRNA- mediated gene silencing in the nucleus. RNA, 2013, 19(1): 17-35.
pmid: 23150874 |
| [66] |
Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell, 2009, 136(3): 496-507.
pmid: 19167051 |
| [67] |
Liu X, Guo JW, Lin XC, Tuo YH, Peng WL, He SY, Li ZQ, Ye YC, Yu J, Zhang FR, Ma MM, Shang JY, Lv XF, Zhou AD, Ouyang Y, Wang C, Pang RP, Sun JX, Ou JS, Zhou JG, Liang SJ. Macrophage NFATc3 prevents foam cell formation and atherosclerosis: evidence and mechanisms. Eur Heart J, 2021, 42(47): 4847-4861.
pmid: 34570211 |
| [68] |
Turunen MP, Lehtola T, Heinonen SE, Assefa GS, Korpisalo P, Girnary R, Glass CK, Väisänen S, Ylä-Herttuala S. Efficient regulation of VEGF expression by promoter-targeted lentiviral shRNAs based on epigenetic mechanism: a novel example of epigenetherapy. Circ Res, 2009, 105(6): 604-609.
pmid: 19696410 |
| [69] |
Liu JH, Liu F. The Yin and Yang function of microRNAs in insulin signalling and cancer. RNA Biol, 2021, 18(1): 24-32.
pmid: 32746694 |
| [70] |
Zhang J, Zhou Y, Wu YJ, Li MJ, Wang RJ, Huang SQ, Gao RR, Ma L, Shi HJ, Zhang J. Hyper-methylated miR-203 dysregulates ABL1 and contributes to the nickel- induced tumorigenesis. Toxicol Lett, 2013, 223(1): 42-51.
pmid: 23968727 |
| [71] |
Zhou MR, Zeng JP, Wang XM, Wang XY, Huang T, Fu Y, Sun T, Jia JH, Chen CY. Histone demethylase RBP2 decreases miR-21 in blast crisis of chronic myeloid leukemia. Oncotarget, 2015, 6(2): 1249-1261.
pmid: 25575817 |
| [72] |
Li CH, Xiao ZG, Tong JHM, To KF, Fang XD, Cheng AS, Chen YC. EZH2 coupled with HOTAIR to silence microRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int J Cancer, 2017, 140(1): 120-129.
pmid: 27594424 |
| [73] |
Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP, Wang F, Sun SH. The histone deacetylase 4/SP1/ microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology, 2011, 54(6): 2025-2035.
pmid: 21837748 |
| [74] |
Dragomir MP, Knutsen E, Calin GA. SnapShot: unconventional miRNA functions. Cell, 2018, 174(4): 1038-1038. e1.
pmid: 30096304 |
| [75] |
Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci USA, 2006, 103(46): 17337-17342.
pmid: 17085592 |
| [76] |
Wang BB, Li SQ, Qi HH, Chowdhury D, Shi Y, Novina CD. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol, 2009, 16(12): 1259-1266.
pmid: 19946268 |
| [77] | Feng Z, Mei SQ, Wu HY, Peng XW, Sun H, Li LH. The research strategy of animal promoter. Journal of Henan Agricultural Sciences, 2012, 41(12): 20-23. |
| 冯政, 梅书棋, 武华玉, 彭先文, 孙华, 李良华. 动物启动子的研究策略. 河南农业科学, 2012, 41(12): 20-23. | |
| [78] | Xiong CJ, Jiang LM. Research progress of enhancer and its bioinformatics prediction. Chemistry of Life, 2011, 31(3): 446-449. |
| 熊春江, 江黎明. 增强子及其生物信息学预测. 生命的化学, 2011, 31(3): 446-449. | |
| [79] | Sun CB, Zhang X. Advance in the research on super- enhancer. Hereditas(Beijing), 2016, 38(12): 1056-1068. |
| 孙长斌, 张曦. 超级增强子研究进展. 遗传, 2016, 38(12): 1056-1068. | |
| [80] |
Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA, 2010, 107(50): 21931-21936.
pmid: 21106759 |
| [81] |
Jin QH, Yu LR, Wang LF, Zhang ZJ, Kasper LH, Lee JE, Wang CC, Brindle PK, Dent SYR, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J, 2011, 30(2): 249-262.
pmid: 21131905 |
| [82] | Yu WQ, Wang YM. Advances research in chromatin DNase I hypersensitive sites and its transcriptional regulation function. J Med Postgra, 2015, 28(6): 666-669. |
| 于文琪, 王雅梅. 染色质DNase I超敏感位点的定位及其转录调控功能的研究进展. 医学研究生学报, 2015, 28(6): 666-669. | |
| [83] |
Wang D, Garcia-Bassets I, Benner C, Li WB, Su X, Zhou YM, Qiu JS, Liu W, Kaikkonen MU, Ohgi KA, Glass CK, Rosenfeld MG, Fu XD. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature, 2011, 474(7351): 390-394.
pmid: 21572438 |
| [84] |
Toscano-Garibay JD, Aquino-Jarquin G. Transcriptional regulation mechanism mediated by miRNA-DNA·DNA triplex structure stabilized by Argonaute. Biochim Biophys Acta, 2014, 1839(11): 1079-1083.
pmid: 25086339 |
| [85] |
Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells, 2012, 17(3): 218-233.
pmid: 22280061 |
| [86] |
Place RF, Wang J, Noonan EJ, Meyers R, Manoharan M, Charisse K, Duncan R, Huang V, Wang XL, Li LC. Formulation of small activating RNA into lipidoid nanoparticles inhibits xenograft prostate tumor growth by inducing p21 expression. Mol Ther Nucleic Acids, 2012, 1(3): e15.
pmid: 23343884 |
| [87] | Li SY, Wan FS. Resaerch progress in miRNA and tumor metastasis. Journal of Nanchang University (Medical Sciences), 2018, 58(3): 77-81. |
| 李淑英, 万福生. miRNA与肿瘤转移的研究进展. 南昌大学学报(医学版), 2018, 58(3): 77-81. |
| [1] | Ning Zhang, Ye Tian. Design strategies and applications of fluorescent protein-based probes [J]. Hereditas(Beijing), 2025, 47(7): 711-728. |
| [2] | Shuting Quan, Weiwei Jiao, Fang Xu, Lin Sun, Hui Qi, Adong Shen. Advances in the regulation of inflammasome activation by GBP family in infectious diseases [J]. Hereditas(Beijing), 2023, 45(11): 1007-1017. |
| [3] | Xinyuan Wang, Yu Zhang, Nan Yang, He Cheng, Yujie Sun. DNMT3a mediates paclitaxel-induced abnormal expression of LINE-1 by increasing the intragenic methylation [J]. Hereditas(Beijing), 2020, 42(1): 100-111. |
| [4] | Ziying Huang, Long Li, Qianqian Li, Xiangdong Liu, Changchun Li. The effect of lncRNA TCONS_00815878 on differentiation of porcine skeletal muscle satellite cells [J]. Hereditas(Beijing), 2019, 41(12): 1119-1128. |
| [5] | Jun Wu, Junhong Zhang, Menghui Huang, Minhui Zhu, Zaikang Tong. Expression analysis of miR164 and its target gene NAC1 in response to low nitrate availability in Betula luminifera [J]. HEREDITAS(Beijing), 2016, 38(2): 155-162. |
| [6] | Xue Zhou, Yilan Du, Ping Jin, Fei Ma. Bioinformatic analysis of cancer-related microRNAs and their target genes [J]. HEREDITAS(Beijing), 2015, 37(9): 855-864. |
| [7] | Yue Li, Xiaodong Liu, Yongmei Dong, Zongming Xie, Shouyi Chen. Cloning and functional analysis of the cotton Trihelix transcription factor GhGT29 [J]. HEREDITAS(Beijing), 2015, 37(12): 1218-1227. |
| [8] | Yan Cheng, Lin Chen, Xin Cao, Siqimeige Ha, Xiaodong Xie. Expression profiling and functional analysis of hsa-miR-125b and its target genes in drug-resistant cell line of human gastric cancer [J]. Hereditas(Beijing), 2014, 36(2): 119-126. |
| [9] | Shaoyin Fu, Hongli Zhao, Zhuqing Zheng, Jinquan Li, Wenguang Zhang. Melatonin regulating the expression of miRNAs involved in hair folli-cle cycle of cashmere goats skin [J]. HEREDITAS(Beijing), 2014, 36(12): 1235-1242. |
| [10] | Chunyan Fan, Qiang Wei, Zhiqiang Hao, Guanglin Li. Prediction and functional analysis of lincRNAs targeted by miRNAs [J]. HEREDITAS(Beijing), 2014, 36(12): 1226-1234. |
| [11] | Guangxin Sun, Yushi Luan, Juanjuan Cui. Mining and characterization of miRNAs closely associated with the pathogenicity in tomato [J]. HEREDITAS, 2014, 36(1): 69-76. |
| [12] | TANG Xiao-Li DENG Li-Bin LIN Jia-Ri ZHANG Wei-Long LIU Shuang-Mei WEI Yi MEI Pu-Ming WANG Yan LIANG Shang-Dong. Sterol regulatory element binding protein 1 and its target gene networks [J]. HEREDITAS, 2013, 35(5): 607-615. |
| [13] | TANG Hai-Ming, CHEN Hong, ZHANG Jing, REN Jing-Yi, XU Ning. Application of next generation sequencing in microRNA detection [J]. HEREDITAS, 2012, 34(6): 784-792. |
| [14] | ZHENG Wei-Hao, LIN Zhi-Qiang, ZHUO Min, DU Hong-Li, WANG Xiao-Ning. Research progress on influenza antiviral small RNAs [J]. HEREDITAS, 2012, 34(5): 526-532. |
| [15] | ZHANG Lei, CHAO Jiang-Chao, CUI Meng-Meng, CHEN Ya-Qiong, ZONG Feng, SUN Yu-Ge. Bioinformatic prediction of conserved microRNAs and their target genes in eggplant (Solanum melongena L.) [J]. HEREDITAS, 2011, 33(7): 776-784. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||