遗传 ›› 2026, Vol. 48 ›› Issue (1): 87-101.doi: 10.16288/j.yczz.25-080
宋凯月1,2( ), 张翼飞1,2(
), 张翼飞1,2( ), 张瑶2, 龙淑婷2, 彭建军2, 孙远东2, 崔小娟2, 潘本山2, 柴语心2, 王晓霞2, 陈家慧2
), 张瑶2, 龙淑婷2, 彭建军2, 孙远东2, 崔小娟2, 潘本山2, 柴语心2, 王晓霞2, 陈家慧2
收稿日期:2025-04-03
修回日期:2025-05-07
出版日期:2026-01-20
发布日期:2025-05-30
通讯作者:
张翼飞,博士,副教授,研究方向:发育生物学。E-mail: zhangyf@hnust.edu.cn作者简介:宋凯月,硕士研究生,专业方向:生物与医药。E-mail: 2628211150@qq.com
基金资助:
Kaiyue Song1,2( ), Yifei Zhang1,2(
), Yifei Zhang1,2( ), Yao Zhang2, Shuting Long2, Jianjun Peng2, Yuandong Sun2, Xiaojuan Cui2, Benshan Pan2, Yuxin Chai2, Xiaoxia Wang2, Jiahui Chen2
), Yao Zhang2, Shuting Long2, Jianjun Peng2, Yuandong Sun2, Xiaojuan Cui2, Benshan Pan2, Yuxin Chai2, Xiaoxia Wang2, Jiahui Chen2
Received:2025-04-03
Revised:2025-05-07
Published:2026-01-20
Online:2025-05-30
Supported by:摘要:
根据“预模式假说(Prepattern Hypothesis)”,黑腹果蝇(Drosophila melanogaster)背部感觉器官前体(sensory organ precursors,SOP)的发育依赖于预模式基因、原神经基因和神经源性基因的依次调控。大多数情况下,单一调控基因的功能失常引起的异位SOP形成,或表现为空间不连续,或仅限于原神经簇内。而fred基因的敲低会造成一种异位SOP发生的新型表型,这些异位SOP在空间上是连续的,且不局限于原神经簇区域。本研究发现,由敲减fred所诱导的异位SOP形成不依赖于原神经簇,表明几乎所有翅成虫盘细胞都具有潜在的神经发生潜力。并且,本研究还发现预模式基因pannier(pnr)与fred协同作用,以两种截然不同的方式调控SOP细胞的命运:一方面,pnr和fred在背板中部对于内源性SOP的形成是必需的;但另一方面,它们在原神经簇外协同抑制SOP发生。本研究结果打破了“预模式假说”中SOP形成依赖于原神经簇形成的论断,为该假说提供了一项重要补充。
宋凯月, 张翼飞, 张瑶, 龙淑婷, 彭建军, 孙远东, 崔小娟, 潘本山, 柴语心, 王晓霞, 陈家慧. pnr与fred协同介导果蝇背板中部SOP细胞发生的双重调控[J]. 遗传, 2026, 48(1): 87-101.
Kaiyue Song, Yifei Zhang, Yao Zhang, Shuting Long, Jianjun Peng, Yuandong Sun, Xiaojuan Cui, Benshan Pan, Yuxin Chai, Xiaoxia Wang, Jiahui Chen. pnr cooperate with fred to regulate SOP cell fate both positively and negatively in the medial notum of Drosophila[J]. Hereditas(Beijing), 2026, 48(1): 87-101.
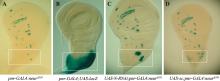
图1
果蝇翅成虫盘中感觉器官前体(SOP)异位发生的两种常见模式 A:基因型为pnr-GAL4:neurA101的翅成虫盘X-gal染色,显示对照成虫盘上正常SOP分布(图片下方位于背板中部的2个SOP);B:pnr>lacZ翅成虫盘的X-gal染色,显示pnr-GAL4的表达区域;C:基因型为pnr>N-RNAi;neurA101的翅成虫盘X-gal染色,显示在背板中部敲低N表达时,异位发生的SOP局限于背板中部两个原神经簇内,呈连续分布;D:基因型为pnr>sc;neurA101的翅成虫盘X-gal染色,显示在背板中部过表达sc时,异位发生的SOP虽可位于两个原神经簇之外,但呈不连续分布。"

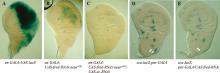
图2
fred基因敲低在原神经簇外诱导异位SOP的产生 A:en>lacZ果蝇翅成虫盘的X-gal染色显示在en-GAL4的表达区域;B、C:基因型为 en>fred-RNAi;neurA101-lacZ(B)和 en>fred-RNAi;sc-RNAi;neurA101-lacZ (C)的翅成虫盘X-gal染色,显示由敲低 fred引发的异位neurA101表达可被sc的敲低所抑制;D、E:基因型为 sca-lacZ;pnr-GAL4(D)和sca-lacZ;pnr>fred-RNAi (E)的翅成虫盘X-gal染色,显示在背板中部fred 敲低背景下,位于该区域的两个原神经簇的形成(白色箭头所示)受到抑制。"
表1
在fred-RNAi微阵列分析中表达上调的基因"
| 基因/别名 | 平均对照信号 | 平均实验信号 | 差异倍数 | 功能/同源性 |
|---|---|---|---|---|
| 转录因子 (6种) | ||||
| Sox100B | 124 | 412 | 3.3 | HMG-box |
| cabut | 201 | 574 | 2.8 | Zinc finger, C2H2 |
| CG6272 | 164 | 413 | 2.5 | bZIP |
| CG18358 | 145 | 353 | 2.4 | LIM domain |
| CG10916 | 291 | 1,541 | 5.3 | Zinc ion binding |
| Arc1 | 117 | 2,642 | 22.6 | Zn-finger CCHC type |
| 酶 (30种) | ||||
| CG18522 | 31 | 670 | 21.0 | Aldehyde oxidase |
| CG3074 | 19.5 | 368 | 18.9 | Endopeptidase |
| CG18547 | 139 | 3,165 | 5.8 | NAD(P)-linked oxidoreductase |
| Nos | 13 | 239 | 17.7 | Nitric-oxide synthase |
| CG2064 | 60 | 645 | 10.8 | Oxidoreductase |
| CG18493 | 41 | 330 | 8.1 | Serine-type peptidase |
| CG3734 | 200 | 1,570 | 7.8 | Serine-type peptidase |
| CG32549 | 139 | 1,041 | 7.5 | 5′-nucleotidase |
| GstE1 | 570 | 3,684 | 6.5 | Glutathione transferase |
| GstE5 | 163 | 930 | 5.7 | Glutathione transferase |
| GstE6 | 70 | 3,051 | 43.6 | Glutathione transferase |
| amd | 130 | 691 | 5.3 | Dopamine metabolism |
| CG1582 | 178 | 692 | 3.9 | Helicase |
| CG12224 | 245 | 852 | 3.5 | Oxidoreductase |
| CG10638 | 1,406 | 4,835 | 3.4 | Aldehyde reductase |
| CG5955 | 140 | 462 | 3.2 | UDP-glucose 4-epimerase |
| CG3008 | 282 | 905 | 3.2 | Protein kinase |
| CG6330 | 104 | 325 | 3.1 | Uridine phosphorylase |
| kraken | 192 | 563 | 2.9 | Serine hydrolase |
| GstD3 | 1,356 | 3,966 | 2.9 | Glutathione transferase |
| CG4210 | 289 | 818 | 2.8 | Protein acetylation |
| Aldh | 445 | 1,238 | 2.8 | Aldehyde dehydrogenase |
| CG31549 | 526 | 1,434 | 2.7 | Oxidoreductase |
| mre11 | 457 | 1,223 | 2.7 | Exonuclease |
| AcCoAS | 116 | 307 | 2.6 | Acetate-CoA ligase |
| Ugt302C1 | 232 | 602 | 2.6 | Glucuronosyltransferase |
| Ugt302K1 | 23 | 429 | 18.5 | UDP-glucosyl transferase |
| MRP | 620 | 1,610 | 2.6 | Xenobiotic-transporting ATPase |
| l(2)03659 | 141 | 362 | 2.6 | ATPase |
| escl | 599 | 1,671 | 2.8 | Histone methyltransferase |
| 结构蛋白 (4种) | ||||
| Tsp42Ed | 19 | 523 | 27.7 | Tetraspanin |
| Ndg | 51 | 335 | 6.6 | Cell adhesion |
| CG3397 | 119 | 529 | 4.4 | Ion-channel |
| Cpr49Ah | 284 | 842 | 3.0 | Structural constituent of cuticle |
| 受体/载体蛋白 (6种) | ||||
| CytC1 | 48 | 372 | 7.8 | Electron transporter |
| glob1 | 120 | 682 | 5.6 | Oxygen transporter |
| Cyp6d2 | 139 | 745 | 5.3 | Electron transporter |
| CG11897 | 109 | 487 | 4.4 | Multidrug transporter |
| slf | 215 | 886 | 4.1 | C-lectin like |
| CG10126 | 134 | 480 | 3.6 | Calcium ion binding |
| 其他蛋白 (27种) | ||||
| JhI-26 | 77 | 348 | 4.5 | Juvenile hormone-inducible protein |
| Thor | 140 | 575 | 4.1 | Immune response |
| bgcn | 920 | 2,469 | 2.7 | RNA-binding protein |
| rev7 | 155 | 550 | 3.5 | Cell cycle regulator |
| upd1 | 233 | 724 | 3.0 | Receptor binding |
| Nplp2 | 219 | 660 | 3.0 | Neuropeptide hormone |
| CG14786 | 123 | 343 | 2.8 | Cell cycle |
| Cul-2 | 467 | 1,298 | 2.8 | Nuclear ubiquitin ligase complex |
| CG8927 | 140 | 374 | 2.7 | Exocytosis |
| CG4858 | 440 | 1,158 | 2.6 | Nucleotide binding |
| ver | 148 | 558 | 3.8 | Telomere capping |
| Ude | 136 | 376 | 2.7 | DNA catabolic process |
| Qtzl | 307 | 803 | 2.6 | Metabolic process |
| vir-1 | 168 | 426 | 2.5 | Defense response to virus |
| CG12640 | 164 | 409 | 2.5 | ATP binding |
| Arc2 | 14 | 723 | 53.4 | |
| CG14059 | 237 | 3,659 | 15.4 | |
| CG15784 | 415 | 4,120 | 9.9 | |
| CG2909 | 110 | 831 | 7.6 | |
| Osi14 | 124 | 602 | 4.8 | |
| TwdlE | 195 | 826 | 4.2 | |
| CG6353 | 230 | 752 | 3.2 | |
| CG16787 | 112 | 356 | 3.2 | |
| CG15347 | 143 | 412 | 2.9 | |
| CG9411 | 38 | 127 | 3.3 | |
| Broadened | 307 | 754 | 2.4 |
表2
在fred-RNAi微阵列分析中表达下调的基因"
| 基因/别名 | 平均对照信号 | 平均实验信号 | 差异倍数 | 功能/同源性 |
|---|---|---|---|---|
| 转录因子 (20种) | ||||
| Doc3 | 294 | 104 | 2.8 | P53 like transcription factor |
| klu | 355 | 132 | 2.7 | Zn finger (C2H2) |
| sr | 316 | 116 | 2.7 | Zn finger (C2H2) |
| dve | 561 | 225 | 2.5 | Homeobox |
| pnr | 284 | 127 | 2.2 | Zn finger (GATA) |
| ap | 1,149 | 564 | 2.0 | Homeobox/LIM domain |
| E(spl)mβ | 3,930 | 1,940 | 2.0 | N signaling pathway |
| psq | 747 | 377 | 2.0 | bHLH |
| salm | 508 | 249 | 2.0 | Transcription factor |
| ara | 596 | 321 | 1.9 | Homeobox |
| vg | 1,250 | 665 | 1.9 | Zn finger (GATA) |
| grn | 341 | 190 | 1.8 | Zn finger (GATA) |
| hth | 1,638 | 932 | 1.8 | Homeobox |
| mirr | 937 | 526 | 1.8 | Homeobox |
| grh | 767 | 453 | 1.7 | bHLH |
| E(spl)m7 | 1,084 | 656 | 1.7 | N signaling pathway |
| E(spl)mδ | 573 | 363 | 1.6 | N signaling pathway |
| E(spl)m3 | 1,901 | 1,183 | 1.6 | N signaling pathway |
| exd | 1,085 | 659 | 1.6 | Homeobox |
| nub | 396 | 167 | 2.4 | POU domain |
| 信号转导蛋白 (2种) | ||||
| E(spl)m2 | 582 | 263 | 2.2 | N signaling pathway |
| E(spl)mα | 1,525 | 722 | 2.1 | N signaling pathway |
| 酶 (15种) | ||||
| Gnpda | 594 | 229 | 2.6 | Glucosamine-6-phosphate deaminase |
| CG4914 | 733 | 288 | 2.5 | Serine endopeptidase |
| CG42514 | 1,633 | 665 | 2.4 | Adenylate cyclase |
| CG16733 | 437 | 186 | 2.3 | Retinol dehydratase |
| fbp | 397 | 170 | 2.3 | Fructose-bisphosphatase |
| Ance | 2,318 | 1,031 | 2.2 | Peptidyl-dipeptidase |
| Fkbp14 | 438 | 210 | 2.1 | Peptidyl-prolyl cis-trans isomerase |
| Spn43Aa | 2,371 | 1,155 | 2.1 | Serine protease inhibitor |
| SAK | 694 | 342 | 2.0 | Serine/threonine kinase |
| tok | 676 | 349 | 2.0 | Procollagen C-endopeptidase |
| RasGAP1 | 253 | 135 | 1.9 | EGFR signaling antagonist |
| CG9416 | 439 | 85 | 5.2 | Peptidase |
| Tpst | 504 | 229 | 2.2 | Protein-tyrosine sulfotransferase |
| Cht3 | 592 | 282 | 2.1 | Chitinase activity |
| Cht1 | 877 | 420 | 2.1 | Chitinase activity |
| 细胞黏附 (2种) | ||||
| CG5758 | 284 | 107 | 2.6 | Beta-Ig-H3/Fasciclin domain |
| leak | 645 | 308 | 2.1 | Immunoglobulin C-2 type |
| 受体/载体蛋白 (7种) | ||||
| CG10960 | 266 | 74 | 3.6 | Glucose transporter |
| Cyp310a1 | 628 | 187 | 3.4 | Electron transporter |
| CG3649 | 241 | 82 | 2.9 | Sodium symporter |
| CG32447 | 248 | 99 | 2.5 | GABA-B-like receptor |
| mnd | 282 | 125 | 2.3 | Amino acid transporter |
| LpR2 | 379 | 180 | 2.1 | Low-density lipoprotein receptor |
| CG1607 | 1,494 | 720 | 2.1 | Amino acid transporter |
| 其他蛋白 (14种) | ||||
| wbl | 573 | 113.4 | 3.6 | Protein processing |
| ImpE3 | 445 | 147 | 3.0 | Imaginal disc eversion |
| bnb | 5,095 | 1,706 | 3.0 | Gliogenesis |
| cpb | 2,227 | 855 | 2.6 | F-actin capping |
| CG3770 | 453 | 199 | 2.3 | Cell polarity |
| RpS12 | 511 | 227 | 2.2 | Structural constituent of ribosome |
| ph-p | 469 | 217 | 2.2 | DNA binding |
| ImpE2 | 10,453 | 4,875 | 2.1 | Imaginal disc eversion |
| Tl | 334 | 162 | 2.1 | Antimicrobial humoral response |
| Lcp2 | 1,408 | 683 | 2.1 | Structural constituent of cuticle |
| bib | 251 | 144 | 1.7 | Connexon channel activity |
| ana | 857 | 501 | 1.7 | Secreted glycoprotein |
| neo | 1,700 | 618 | 2.8 | Regulation of embryonic cell shape |
| CG15905 | 629 | 267 | 2.4 |
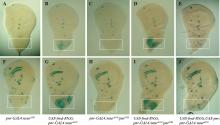
图6
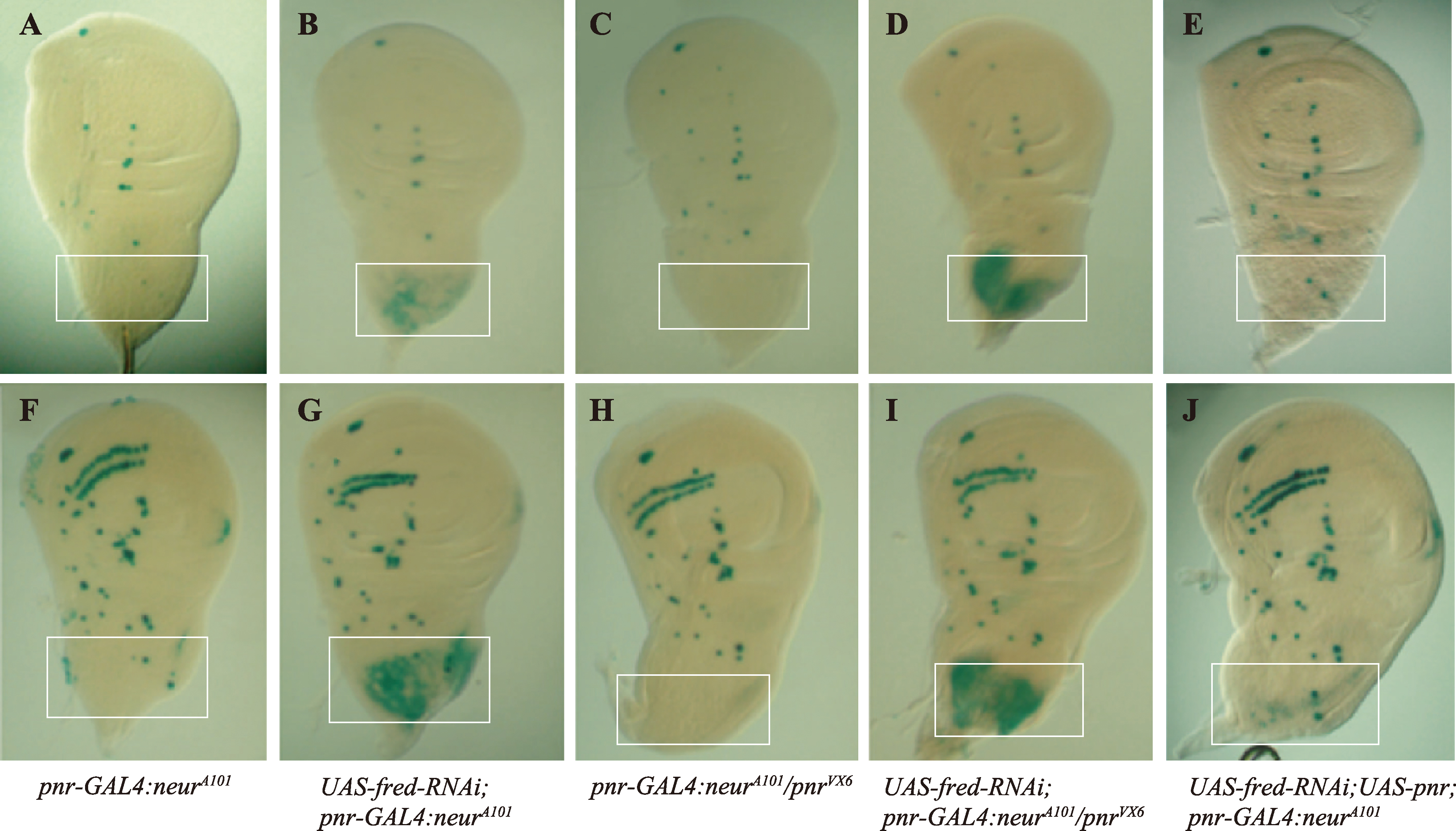
pnr与fred协同抑制原神经簇外SOP发生 A~E、F~J:分别展示了早期三龄和后期三龄幼虫翅成虫盘的neurA101 X-gal染色,以标记SOP的生成情况。基因型为pnr-GAL4:neurA101/+的对照翅成虫盘显示正常的SOP模式(A和 F);用pnr-GAL4驱动敲低fred时,发现SOP在背板中部出现了异位表达(B和G);在pnrVX6/pnr-GAL4:neurA101翅成虫盘中,背板中部的SOP缺失(C和H);当在pnrVX6/pnr-GAL4背景中敲低fred 时,异位SOP表型显著增强(D和I);而在背板中部过表达pnr则几乎完全抑制了由敲低fred引起的异位SOP表型(E和J)。白边方框表示位于背板中部的pnr表达区域。"
| [1] |
Koto A, Kuranaga E, Miura M. Apoptosis ensures spacing pattern formation of Drosophila sensory organs. Curr Biol, 2011, 21(4): 278-287.
pmid: 21276725 |
| [2] |
Marcellini S, Simpson P. Two or four bristles: functional evolution of an enhancer of scute in Drosophilidae. PLoS Biol, 2006, 4(12): e386.
pmid: 17105353 |
| [3] |
Emery G, Hutterer A, Berdnik D, Mayer B, Wirtz-Peitz F, Gaitan MG, Knoblich JA. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell, 2005, 122(5): 763-773.
pmid: 16137758 |
| [4] | Troost T, Binshtok U, Sprinzak D, Klein T. Cis-inhibition suppresses basal Notch signalling during sensory organ precursor selection. bioRxiv, 2022, doi: 10.1101/2022.07.19.500683. |
| [5] |
Ghysen A, Dambly-Chaudière C. From DNA to form: the achaete-scute complex. Genes Dev, 1988, 2(5): 495-501.
pmid: 3290049 |
| [6] |
Gómez-Skarmeta JL, Campuzano S, Modolell J. Half a century of neural prepatterning: the story of a few bristles and many genes. Nat Rev Neurosci, 2003, 4(7): 587-598.
pmid: 12838333 |
| [7] |
Ruiz-Gómez M, Modolell J. Deletion analysis of the achaete-scute locus of Drosophila melanogaster. Genes Dev, 1987, 1(10): 1238-1246.
pmid: 3123318 |
| [8] |
Usui K, Goldstone C, Gibert JM, Simpson P. Redundant mechanisms mediate bristle patterning on the Drosophila thorax. Proc Natl Acad Sci USA, 2008, 105(51): 20112-20117.
pmid: 19104061 |
| [9] |
Campuzano S, Modolell J. Patterning of the Drosophila nervous system: the achaete-scute gene complex. Trends Genet, 1992, 8(6): 202-208.
pmid: 1496555 |
| [10] |
Garrell J, Campuzano S. The helix-loop-helix domain: a common motif for bristles, muscles and sex. Bioessays, 1991, 13(10): 493-498.
pmid: 1755826 |
| [11] |
Jan YN, Jan LY. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell, 1993, 75(5): 827-830.
pmid: 8252617 |
| [12] |
He QY, Hou TL, Fan CX, Wang SC, Wang YH, Chen SS. Juvenile hormone suppresses sensory organ precursor determination to block Drosophila adult abdomen morphogenesis. Insect Biochem Mol Biol, 2023, 157: 103957.
pmid: 37192726 |
| [13] |
Dutriaux A, Godart A, Brachet A, Silber J. The insulin receptor is required for the development of the Drosophila peripheral nervous system. Plos One, 2013, 8(9): e71857.
pmid: 24069139 |
| [14] |
Marcellini S, Gibert JM, Simpson P. achaete, but not scute, is dispensable for the peripheral nervous system of Drosophila. Dev Biol, 2005, 285(2): 545-553.
pmid: 16084506 |
| [15] |
Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science, 1999, 284(5415): 770-776.
pmid: 10221902 |
| [16] |
Culí J, Modolell J. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev, 1998, 12(13): 2036-2047.
pmid: 9649507 |
| [17] |
Huppert SS, Jacobsen TL, Muskavitch MA. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development, 1997, 124(17): 3283-3291.
pmid: 9310323 |
| [18] |
Francis C, Lydie C, Hervé R, Khalil M, François S. Self-organized Notch dynamics generate stereotyped sensory organ patterns in Drosophila. Science, 2017, 356(6337): eaai7407.
pmid: 28386027 |
| [19] |
Campos-Ortega JA. Mechanisms of early neurogenesis in Drosophila melanogaster. J Neurobiol, 1993, 24(10): 1305-1327.
pmid: 8228961 |
| [20] |
Miller SW, Posakony JW. Lateral inhibition: two modes of non-autonomous negative autoregulation by neuralized. PLoS Genet, 2018, 14(7): e1007528.
pmid: 30028887 |
| [21] |
Troost T, Binshtok U, Sprinzak D, Klein T. Cis-inhibition suppresses basal Notch signaling during sensory organ precursor selection. Proc Natl Acad Sci USA, 2023, 120(23): e2214535120.
pmid: 37252950 |
| [22] |
Cubas P, Modolell J. The extramacrochaetae gene provides information for sensory organ patterning. EMBO J, 1992, 11(9): 3385-3393.
pmid: 1505522 |
| [23] |
del Corral RD, Aroca P, Gómez-Skarmeta JL, Cavodeassi F, Modolell J. The Iroquois homeodomain proteins are required to specify body wall identity in Drosophila. Genes Dev, 1999, 13(13): 1754-1761.
pmid: 10398687 |
| [24] |
Heitzler P, Haenlin M, Ramain P, Calleja M, Simpson P. A genetic analysis of pannier, a gene necessary for viability of dorsal tissues and bristle positioning in Drosophila. Genetics, 1996, 143(3): 1271-1286.
pmid: 8807299 |
| [25] |
Tobias T, Markus S, Thomas K. A re-examination of the selection of the sensory organ precursor of the bristle sensilla of Drosophila melanogaster. PLoS Genet, 2015, 11(1): e1004911.
pmid: 25569355 |
| [26] |
Chandra S, Ahmed A, Vaessin H. The Drosophila IgC2 domain protein Friend-of-Echinoid, a paralogue of Echinoid, limits the number of sensory organ precursors in the wing disc and interacts with the Notch signaling pathway. Dev Biol, 2003, 256(2): 302-316.
pmid: 12679104 |
| [27] |
Fetting JL, Spencer SA, Wolff T. The cell adhesion molecules Echinoid and Friend of Echinoid coordinate cell adhesion and cell signaling to regulate the fidelity of ommatidial rotation in the Drosophila eye. Development, 2009, 136(19): 3323-3333.
pmid: 19736327 |
| [28] |
Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development, 1996, 122(1): 161-171.
pmid: 8565827 |
| [29] |
Calleja M, Moreno E, Pelaz S, Morata G. Visualization of gene expression in living adult Drosophila. Science, 1996, 274(5285): 252-255.
pmid: 8824191 |
| [30] |
Huang F, Dambly-Chaudière C, Ghysen A. The emergence of sense organs in the wing disc of Drosophila. Development, 1991, 111(4): 1087-1095.
pmid: 1879352 |
| [31] |
Ramain P, Heitzler P, Haenlin M, Simpson P. Pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development, 1993, 119(4): 1277-1291.
pmid: 7916679 |
| [32] |
Volk T, VijayRaghavan K. A central role for epidermal segment border cells in the induction of muscle patterning in the Drosophila embryo. Development, 1994, 120(1): 59-70.
pmid: 8119132 |
| [33] |
Ellis MC, Weber U, Wiersdorff V, Mlodzik M. Confrontation of scabrous expressing and non-expressing cells is essential for normal ommatidial spacing in the Drosophila eye. Development, 1994, 120(7): 1959-1969.
pmid: 7925001 |
| [34] |
Haenlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y, Simpson P, Ramain P. Transcriptional activity of pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev, 1997, 11(22): 3096-3108.
pmid: 9367990 |
| [35] |
Cubadda Y, Heitzler P, Ray R P, Bourouis M, Ramain P, Gelbart W, Simpson P, Haenlin M. u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev, 1997, 11(22): 3083-3095.
pmid: 9367989 |
| [36] |
Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development, 1993, 117(4): 1223-1237.
pmid: 8404527 |
| [37] |
Kaspar M, Schneider M, Chia W, Klein T. Klumpfuss is involved in the determination of sensory organ precursors in Drosophila. Dev Biol, 2008, 324(2): 177-191.
pmid: 18831969 |
| [38] |
Mlodzik M, Baker NE, Rubin GM. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev, 1990, 4(11): 1848-1861.
pmid: 2125959 |
| [39] |
Furman DP, Bukharina TA. Morphogenesis of Drosophila melanogaster macrochaetes: cell fate determination for bristle organ. J Stem Cells, 2012, 7(1): 19-41.
pmid: 23550342 |
| [40] |
Bruelle C, Pinot M, Daniel E, Daudé M, Mathieu J, Le Borgne R. Cell-intrinsic and -extrinsic roles of the ESCRT-III subunit Shrub in abscission of Drosophila sensory organ precursors. Development, 2023, 150(10): dev201409.
pmid: 37226981 |
| [41] |
Hainaut M, Sagnier T, Bérenger H, Pradel J, Graba Y, Miotto B. The MYST-containing protein Chameau is required for proper sensory organ specification during Drosophila thorax morphogenesis. PLoS One, 2012, 7(3): e32882.
pmid: 22412942 |
| [42] |
Asmar J, Biryukova I, Heitzler P. Drosophila dLMO-PA isoform acts as an early activator of achaete/scute proneural expression. Dev Biol, 2008, 316(2): 487-497.
pmid: 18329012 |
| [43] |
Tian RB, Chen X, Wu MM, Xu QX, Wang S, Zang LS, Xiao D. The molecular properties and roles of pannier in Harmonia axyridis’s metamorphosis and melanin synthesis. Front Physiol, 2022, 13: 909258.
pmid: 35592031 |
| [44] |
Liang Q, Peng TT, Sun BY, Tu JB, Cheng XY, Tian YLZ, Fan XL, Yang DY, Gaur U, Yang MY. Gene expression patterns determine the differential numbers of dorsocentral macrochaetes between Musca domestica and Drosophila melanogaster. Genesis, 2018, 56(11-12): e23258.
pmid: 30358076 |
| [45] |
García-García MJ, Ramain P, Simpson P, Modolell J. Different contributions of pannier and wingless to the patterning of the dorsal mesothorax of Drosophila. Development, 1999, 126(16): 3523-3532.
pmid: 10409499 |
| [46] |
Fromental-Ramain C, Taquet N, Ramain P. Transcriptional interactions between the pannier isoforms and the cofactor U-shaped during neural development in Drosophila. Mech Dev, 2010, 127(9-12): 442-457.
pmid: 20709169 |
| [47] |
Fromental-Ramain C, Vanolst L, Delaporte C, Ramain P. pannier encodes two structurally related isoforms that are differentially expressed during Drosophila development and display distinct functions during thorax patterning. Mech Dev, 2008, 125(1-2): 43-57.
pmid: 18042352 |
| [48] |
Svetlana M, William T, Ruth S. JAK/STAT and the GATA factor Pannier control hemocyte maturation and differentiation in Drosophila. Dev Biol, 2011, 352(2): 308-316.
pmid: 21295568 |
| [49] |
Frommer G, Vorbrüggen G, Pasca G, Jäckle H, Volk T. Epidermal egr-like zinc finger protein of Drosophila participates in myotube guidance. EMBO J, 1996, 15(7): 1642-1649.
pmid: 8612588 |
| [50] |
Lee JC, VijayRaghavan K, Celniker SE, Tanouye MA. Identification of a Drosophila muscle development gene with structural homology to mammalian early growth response transcription factors. Proc Natl Acad Sci USA, 1995, 92(22): 10344-10348.
pmid: 7479781 |
| [51] |
Ghazi A, Paul L, VijayRaghavan K. Prepattern genes and signaling molecules regulate stripe expression to specify Drosophila flight muscle attachment sites. Mech Dev, 2003, 120(5): 519-528.
pmid: 12782269 |
| [52] |
Usui K, Pistillo D, Simpson P. Mutual exclusion of sensory bristles and tendons on the notum of dipteran flies. Curr Biol, 2004, 14(12): 1047-1055.
pmid: 15202997 |
| [53] |
Amit S, Meghana T, Oorvashi Roy P, Madhuri KS. A glimpse into dorso-ventral patterning of the Drosophila eye. Dev Dyn, 2012, 241(1): 69-84.
pmid: 22034010 |
| [54] |
Gómez-Skarmeta JL, del Corral RD, de la Calle-Mustienes E, Ferrés-Marcó D, Modolell J. araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell, 1996, 85(1): 95-105.
pmid: 8620542 |
| [55] |
Kehl BT, Cho KO, Choi KW. mirror, a Drosophila homeobox gene in the iroquois complex, is required for sensory organ and alula formation. Development, 1998, 125(7): 1217-1227.
pmid: 9477320 |
| [1] | 何江平, 陈捷凯. 转座元件、表观遗传调控与细胞命运决定[J]. 遗传, 2021, 43(9): 822-834. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
