遗传 ›› 2026, Vol. 48 ›› Issue (1): 102-115.doi: 10.16288/j.yczz.25-173
• 研究报告 • 上一篇
郑腾飞( ), 梁歆悦, 孟迎迎, 安亚龙, 王育禾, 史新娥, 李晓(
), 梁歆悦, 孟迎迎, 安亚龙, 王育禾, 史新娥, 李晓( )
)
收稿日期:2025-06-11
修回日期:2025-09-18
出版日期:2026-01-20
发布日期:2025-10-14
通讯作者:
李晓,博士,副教授,研究方向:猪遗传育种研究。E-mail: nicelixiao@nwafu.edu.cn作者简介:郑腾飞,硕士研究生,专业方向:动物遗传育种与繁殖。E-mail: ztf455224189@outlook.com
基金资助:
Tengfei Zheng( ), Xinyue Liang, Yingying Meng, Yalong An, Yuhe Wang, Xin’e Shi, Xiao Li(
), Xinyue Liang, Yingying Meng, Yalong An, Yuhe Wang, Xin’e Shi, Xiao Li( )
)
Received:2025-06-11
Revised:2025-09-18
Published:2026-01-20
Online:2025-10-14
Supported by:摘要:
谷胱甘肽过氧化物酶8 (glutathione peroxidase 8,GPX8)是谷胱甘肽过氧化物酶家族的重要成员。全基因组关联分析(genome-wide association analysis,GWAS)表明,GPX8与猪生长发育及胴体性状高度相关。本研究以猪骨骼肌卫星细胞为研究对象,分别干扰和过表达GPX8,利用免疫荧光染色、qRT-PCR和Western blotting等方法探究GPX8对成肌分化和肌纤维类型转化的影响。结果显示,干扰GPX8显著提高成肌分化指数(P<0.01),促进分化标志基因MyHC、MyoG mRNA和蛋白的表达(P<0.05),过表达GPX8得到相反的结果。干扰GPX8显著降低MYH7 mRNA(P<0.01)和慢肌纤维蛋白(slow-twitch MyHC,slow-MyHC)的表达(P<0.05),抑制线粒体生物发生,过表达GPX8得到相反的结果。整合GWAS等多组学数据筛选出调控GPX8表达的顺式eQTLs (expression quantitative trait locus),并通过双荧光素酶报告分析检测候选SNPs对GPX8启动子活性的影响,鉴定出GPX8启动子区5个候选SNPs (rs335618489、rs325233940、rs32989756、rs322106839和rs701033890),其中rs335618489-T、rs325233940-G、rs32989756-T和rs322106839-G通过改变启动子活性显著上调GPX8表达水平(P<0.01),进而影响猪产肉性状。总之,本研究表明GPX8抑制猪骨骼肌卫星细胞成肌分化并促进快肌纤维向慢肌纤维类型转化,GPX8启动子区功能性SNPs通过调控GPX8表达影响猪肌肉发育,为进一步提高猪肉产量提供了育种靶点。
郑腾飞, 梁歆悦, 孟迎迎, 安亚龙, 王育禾, 史新娥, 李晓. GPX8抑制猪骨骼肌卫星细胞成肌分化并促进慢肌纤维形成[J]. 遗传, 2026, 48(1): 102-115.
Tengfei Zheng, Xinyue Liang, Yingying Meng, Yalong An, Yuhe Wang, Xin’e Shi, Xiao Li. GPX8 inhibits myogenic differentiation and promotes slow myofiber formation of porcine skeletal muscle satellite cells[J]. Hereditas(Beijing), 2026, 48(1): 102-115.
表1
野生型及突变型载体的氨基酸序列"
| 野生型 | 突变型 |
|---|---|
| MEPLTAYPLR CSGPKAKVFA VLLSMVLCTV MLFLLQLKFL KPKINSFYNF EVKDAKGRTV SLEKFKGKVA LVVNVASDCQ LTDRNYLALQ ELHKEFGPFH FSVLAFPCNQ FGESEPRPSK EVLSFARNNY GVTFPIFHKI KILGSEAEPA FRFLVDSSKK EPRWNFWKYL VNPEGQVVKY WRPEEPIEVI RPEIAALIRP MIIKKKEDL | MEPLTAYPLR CSGPKAKVFA VLLSMVLATV MLFLLQLKFL KPKINSFYNF EVKDAKGRTV SLEKFKGKVA LVVNVASDSQ LTDRNYLALQ ELHKEFGPFH FSVLAFPCNQ FGESEPRPSK EVLSFARNNY GVTFPIFHKI KILGSEAEPA FRFLVDSSKK EPRWNFWKYL VNPEGQVVKY WRPEEPIEVI RPEIAALIRP MIIKKKEDL |
表2
实时定量PCR引物序列"
| 基因 | 引物序列(5'→3') |
|---|---|
| MyoD | F:CGGCTCTCTCTGCTCCTTTG R:GTCGAAACACGGGTCATCA |
| MyoG | F:GACCCTACAGACGCCCACAA R:CCGTGATGCTGTCCACGAT |
| MyHC | F:CGCAAGAATGTTCTCAGGCT R:GCCAGGTTGACATTGGATTG |
| MYH7 | F:AAGGGCTTGAACGAGGAGTAGA R:TTATTCTGCTTCCTCCAAAGGG |
| MYH2 | F:AAGTGACTGTGAAAACAGAAGCA R:GCAGCCATTTGTAAGGGTTGAC |
| MYH4 | F:AAACCACCTCAGAGTTGTGGA R:GTTCCGAAGGTTCCTGATTGC |
| MYH1 | F:ACAACCCCTACGATTATGCGT R:TTTATGGTCCGTGTGGGTCC |
| MT-ND1 | F:AATCGCCCTTCTCCTTCCCCTAC R:TGGGTTCATTCGTATGCTAGGCTTG |
| MT-ND2 | F:CCTTCACCGCCACCGAACTAATC R:TCTGTTTGGTTTCCTCAGCGTGTG |
| MT-COX1 | F:ACAAAGACATCGGCACCCT R:GATCATCGCCAAGTAGGGTT |
| MT-COX2 | F:TCCCAGGACGACTAAACCAAACAAC R:AGTACAATGGGCATGAAGCTGTGG |
| MT-COX3 | F:CATTCTGAGGAGCTACGGTCATCAC R:GGCAGGATAAAGTGGAAGGCGAAG |
| GPX8 | F:AAGCAAAGCACTGCAGGAAC R:CCCTCAGGGTTGACCAGATAC |
| β-actin | F:TGCTGAGTATGTCGTGGAGTCT R:ATGCATTGCTGACAATCTTGAG |
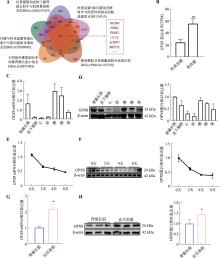
图1
GPX8在猪不同组织和骨骼肌卫星细胞分化过程中的表达模式 A:5个与成肌分化或猪肉质相关转录组数据综合分析;B:GPX8在杜洛克猪和荣昌猪背最长肌中的表达情况(TPM表示每百万转录本数);C:GPX8 mRNA在猪不同组织中的表达情况;D:GPX8蛋白在猪不同组织中的表达情况及量化统计结果;E:GPX8 mRNA在诱导分化0天、2天、4天、8天的猪骨骼肌卫星细胞中的表达情况;F:GPX8蛋白在成肌分化0天、2天、4天、8天的表达情况及量化统计结果;G:GPX8 mRNA在背最长肌和比目鱼肌中的表达情况;H:GPX8蛋白在背最长肌和比目鱼肌中的表达情况及量化统计结果。n=3;*P<0.05,**P<0.01。"
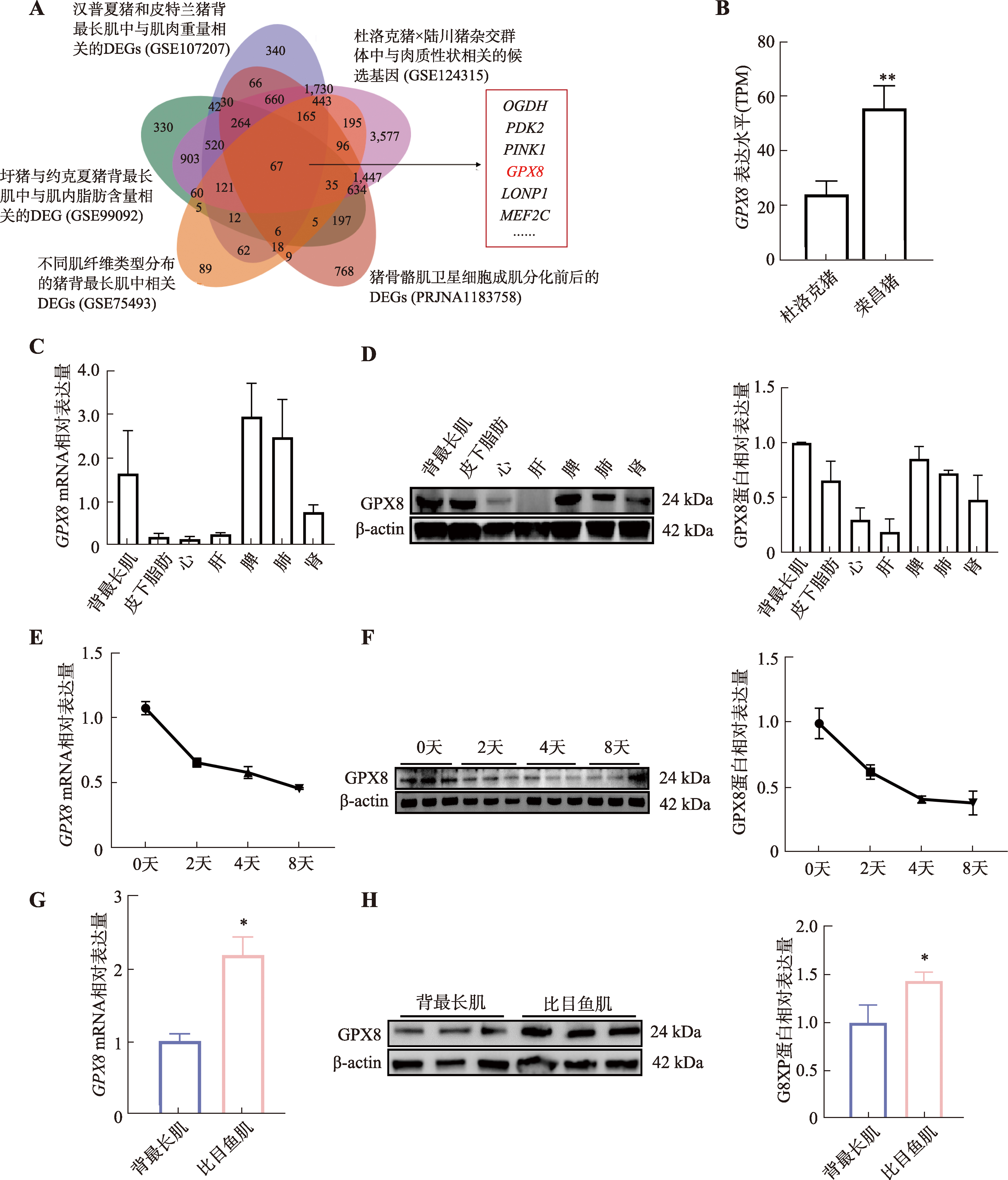
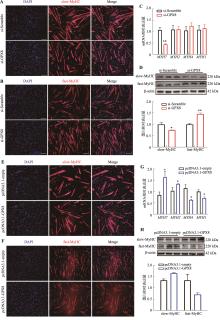
图3
GPX8促进慢肌纤维生成 A:干扰GPX8后慢肌纤维蛋白(slow-MyHC)免疫荧光染色结果;B:干扰GPX8后快肌纤维蛋白(fast-MyHC)免疫荧光染色结果;C:干扰GPX8后MYHC亚型基因(MYH7、MYH2、MYH4和MYH1)mRNA表达水平;D:干扰GPX8后,slow-MyHC和fast-MyHC表达情况及统计结果;E:过表达GPX8后slow-MyHC的免疫荧光染色结果;F:过表达GPX8后fast-MyHC的免疫荧光染色结果;G:过表达GPX8后MYHC亚型基因(MYH7、MYH2、MYH4和MYH1)mRNA表达水平;H:过表达GPX8后,slow-MyHC和fast-MyHC表达情况及统计结果。n=3;*P<0.05,**P<0.01;比例尺:200 μm。"

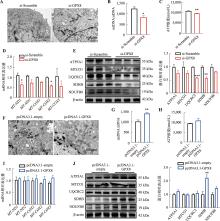
图4
GPX8促进线粒体氧化代谢 A:干扰GPX8后线粒体的透射电镜结果;B:干扰GPX8后mtDNA拷贝数变化;C:干扰GPX8后ATP相对水平变化;D:干扰GPX8后线粒体相关基因mRNA表达情况;E:干扰GPX8后线粒体电子呼吸链相关蛋白的表达情况及统计结果;F:过表达GPX8后线粒体的透射电镜结果;G:过表达GPX8后mtDNA拷贝数变化;H:过表达GPX8后ATP相对水平变化;I:过表达GPX8后线粒体相关基因mRNA表达情况;J:过表达GPX8后线粒体电子呼吸链相关蛋白表达情况及统计结果。n=3;*P<0.05,**P<0.01,***P<0.001;比例尺:500 nm。"

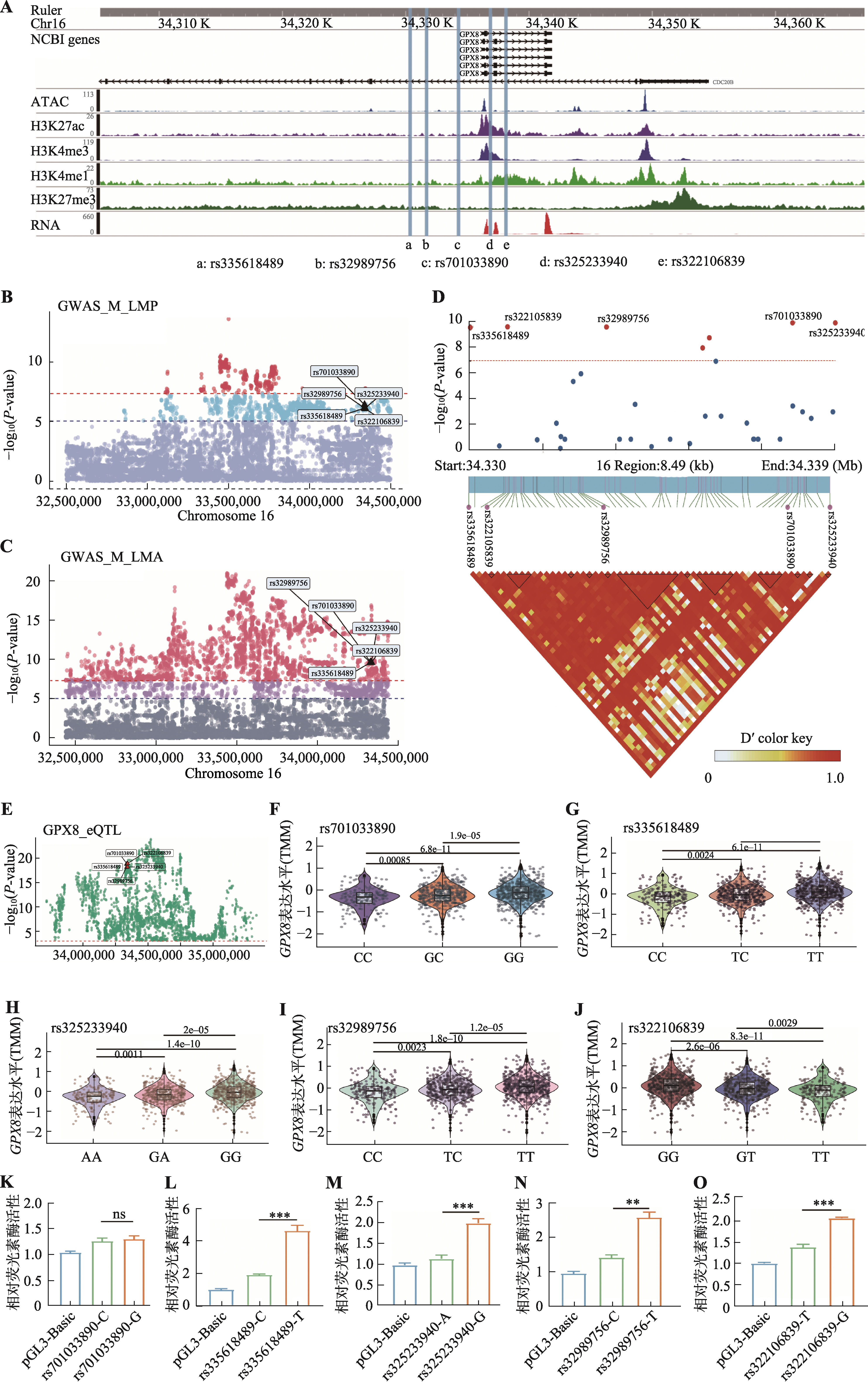
图5
GPX8-eQTL对应的遗传变异通过改变启动子活性调控其表达 A:通过GWAS鉴定猪GPX8基因启动子区域的SNPs;B:通过GWAS表明rs335618489、rs32989756、rs701033890、rs325233940和rs322106839与猪瘦肉率(LMP)显著相关;C:通过GWAS表明rs335618489、rs32989756、rs701033890、rs325233940和rs322106839与猪眼肌面积(LMA)显著相关;D:曼哈顿图(Manhattan plot)和连锁不平衡图(linkage disequilibrium plot)表明rs335618489、rs32989756、rs701033890、rs325233940和rs322106839与LMA和LMP关联性,SNPs位点间存在高度连锁不平衡;E:通过eQTL整合分析,SNPs位点与肌肉组织中GPX8基因表达显著相关;F~J:rs701033890(F)、rs335618489(G)、rs325233940(H)、rs32989756(I)和rs322106839(J)位点突变不同基因型中GPX8表达量的变化;K~O:双荧光素酶报告分析检测rs701033890-C/G(K)、rs335618489-C/T(L)、rs325233940-A/G(M)、rs32989756-C/T(N)和rs322106839-T/G(O)突变前后启动子的活性变化。使用R包edgeR中的TMM(Trimmed Mean of M-values)方法对基因表达量进行样本间标准化;数据以均值±标准误表示(n=3);*P<0.05,**P<0.01,***P<0.001。"
| [1] |
Matarneh SK, Silva SL, Gerrard DE. New insights in muscle biology that alter meat quality. Annu Rev Anim Biosci, 2021, 9: 355-377.
pmid: 33338390 |
| [2] |
Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev, 2011, 91(4): 1447-1531.
pmid: 22013216 |
| [3] |
Wigmore PM, Stickland NC. Muscle development in large and small pig fetuses. J Anat, 1983, 137(Pt 2): 235-245.
pmid: 6630038 |
| [4] |
Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J, 2013, 280(17): 4294-4314.
pmid: 23517348 |
| [5] |
Yang YL, Yan JY, Fan XH, Chen JX, Wang ZS, Liu XQ, Yi GQ, Liu YW, Niu YC, Zhang LC, Wang LX, Li SC, Li K, Tang ZL. The genome variation and developmental transcriptome maps reveal genetic differentiation of skeletal muscle in pigs. PLoS Genet, 2021, 17(11): e1009910.
pmid: 34780471 |
| [6] |
Wang M, Yu H, Kim YS, Bidwell CA, Kuang S. Myostatin facilitates slow and inhibits fast myosin heavy chain expression during myogenic differentiation. BBRC, 2012, 426(1): 83-88.
pmid: 22910409 |
| [7] |
Reisman EG, Caruana NJ, Bishop DJ. Exercise training and changes in skeletal muscle mitochondrial proteins: from blots to “omics”. Crit Rev Biochem Mol Biol, 2024, 59(3-4): 221-243.
pmid: 39288086 |
| [8] |
Sousa-Victor P, García-Prat L, Muñoz-Cánoves P. Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat Rev Mol Cell Biol, 2022, 23(3): 204-226.
pmid: 34663964 |
| [9] |
Choi S, Ferrari G, Tedesco FS. Cellular dynamics of myogenic cell migration: molecular mechanisms and implications for skeletal muscle cell therapies. EMBO Mol Med, 2020, 12(12): e12357.
pmid: 33210465 |
| [10] |
Hernández-Hernández JM, García-González EG, Brun CE, Rudnicki MA. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin Cell Dev Biol, 2017, 72: 10-18.
pmid: 29127045 |
| [11] |
Yartseva V, Goldstein LD, Rodman J, Kates L, Chen MZ, Chen YJJ, Foreman O, Siebel CW, Modrusan Z, Peterson AS, Jovičić A. Heterogeneity of satellite cells implicates DELTA1/NOTCH2 signaling in self-renewal. Cell Rep, 2020, 30(5): 1491-1503.e6.
pmid: 32023464 |
| [12] |
Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta, 2011, 1813(7): 1269-1278.
pmid: 20933024 |
| [13] |
Zhuang ZW, Li SY, Ding RR, Yang M, Zheng EQ, Yang HQ, Gu T, Xu Z, Cai GY, Wu ZF, Yang J. Meta-analysis of genome-wide association studies for loin muscle area and loin muscle depth in two Duroc pig populations. PLoS One, 2019, 14(6): e0218263.
pmid: 31188900 |
| [14] |
Pei J, Pan XY, Wei GH, Hua Y. Research progress of glutathione peroxidase family(GPX) in redoxidation. Front Pharmacol, 2023, 14: 1147414.
pmid: 36937839 |
| [15] |
Kanemura S, Sofia EF, Hirai N, Okumura M, Kadokura H, Inaba K. Characterization of the endoplasmic reticulum- resident peroxidases GPx7 and GPx8 shows the higher oxidative activity of GPx7 and its linkage to oxidative protein folding. J Biol Chem, 2020, 295(36): 12772-12785.
pmid: 32719007 |
| [16] |
Yoboue ED, Rimessi A, Anelli T, Pinton P, Sitia R. Regulation of calcium fluxes by GPX8, a Type-II transmembrane peroxidase enriched at the mitochondria- associated endoplasmic reticulum membrane. Antioxid Redox Signal, 2017, 27(9): 583-595.
pmid: 28129698 |
| [17] |
Yin X, Zhang P, Xia N, Wu SQ, Liu BY, Weng L, Shang MY. GPx8 regulates apoptosis and autophagy in esophageal squamous cell carcinoma through the IRE1/JNK pathway. Cell Signal, 2022, 93: 110307.
pmid: 35288240 |
| [18] |
Zhang J, Liu Y, Guo Y, Zhao Q. GPX8 promotes migration and invasion by regulating epithelial characteristics in non-small cell lung cancer. Thorac Cancer, 2020, 11(11): 3299-3308.
pmid: 32975378 |
| [19] |
Nguyen TTM, Nguyen TH, Kim HS, Dao TTP, Moon Y, Seo M, Kang SM, Mai VH, An YJ, Jung CR, Kim JM, Park S. GPX8 regulates clear cell renal cell carcinoma tumorigenesis through promoting lipogenesis by NNMT. J Exp Clin Cancer Res, 2023, 42(1): 42.
pmid: 36750850 |
| [20] |
Chen H, Xu L, Shan ZL, Chen S, Hu H. GPX8 is transcriptionally regulated by FOXC1 and promotes the growth of gastric cancer cells through activating the Wnt signaling pathway. Cancer Cell Int, 2020, 20(1): 596.
pmid: 33317536 |
| [21] |
Steibel JP, Bates RO, Rosa GJM, Tempelman RJ, Rilington VD, Ragavendran A, Raney NE, Ramos AM, Cardoso FF, Edwards DB, Ernst CW. Genome-wide linkage analysis of global gene expression in loin muscle tissue identifies candidate genes in pigs. PLoS One, 2011, 6(2): e16766.
pmid: 21346809 |
| [22] |
Wang ZZ, Ma HR, Xu L, Zhu B, Liu Y, Bordbar F, Chen Y, Zhang LP, Gao X, Gao HJ, Zhang SL, Xu LY, Li JY. Genome-wide scan identifies selection signatures in Chinese Wagyu cattle using a high-density SNP array. Animals (Basel), 2019, 9(6): 296.
pmid: 31151238 |
| [23] |
Dou ML, Yao Y, Ma L, Wang XY, Shi XE, Yang GS, Li X. The long noncoding RNA MyHC IIA/X-AS contributes to skeletal muscle myogenesis and maintains the fast fiber phenotype. J Biol Chem, 2020, 295(15): 4937-4949.
pmid: 32152230 |
| [24] |
Xu JG, Wang CL, Jin EH, Gu YF, Li SH, Li QG. Identification of differentially expressed genes in longissimus dorsi muscle between Wei and Yorkshire pigs using RNA sequencing. Genes Genomics, 2018, 40(4): 413-421.
pmid: 29892843 |
| [25] |
Ropka-Molik K, Pawlina-Tyszko K, Żukowski K, Piórkowska K, Żak G, Gurgul A, Derebecka N, Wesoły J. Examining the genetic background of porcine muscle growth and development based on transcriptome and mirnaome data. Int J Mol Sci, 2018, 19(4): 1208.
pmid: 29659518 |
| [26] |
Liu Y, Liu XL, Zheng ZW, Ma TT, Liu Y, Long H, Cheng HJ, Fang M, Gong J, Li XY, Zhao SH, Xu XW. Genome- wide analysis of expression QTL (eQTL) and allele- specific expression (ASE) in pig muscle identifies candidate genes for meat quality traits. Genet Sel Evol, 2020, 52(1): 59.
pmid: 33036552 |
| [27] |
Ropka-Molik K, Bereta A, Żukowski K, Piórkowska K, Gurgul A, Żak G. Transcriptomic gene profiling of porcine muscle tissue depending on histological properties. Anim Sci J, 2017, 88(8): 1178-1188.
pmid: 28026080 |
| [28] | An YL, Zhang C, Ge ZH, Li Y, Wen CL, Ding RR, Han PY, Yue YQ, Wu JW, Jin JJ, Li X. The atlas of promoter- enhancer interactions during myogenic differentiation offers novel insights into genetic variants related to meat traits in pigs. J Integr Agric, 2025, doi: 10.1016/j.jia.2025.05.009. |
| [29] |
Fu YH, Liu H, Dou JW, Wang Y, Liao Y, Huang X, Tang ZS, Xu JY, Yin D, Zhu SL, Liu YF, Shen X, Liu HY, Liu JQ, Yang X, Zhang Y, Xiang Y, Li JJ, Zheng ZQ, Zhao YX, Ma YL, Wang HY, Du XY, Xie SS, Xu XW, Zhang HH, Yin LL, Zhu MJ, Yu M, Li XY, Liu XL, Zhao SH. IAnimal: a cross-species omics knowledgebase for animals. Nucleic Acids Res, 2023, 51(D1): D1312-D1324.
pmid: 36300629 |
| [30] |
Dong SS, He WM, Ji JJ, Zhang C, Guo Y, Yang TL. LDBlockShow: a fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief Bioinform, 2021, 22(4): bbaa227.
pmid: 33126247 |
| [31] |
Pan ZY, Yao YL, Yin HW, Cai ZX, Wang Y, Bai LJ, Kern C, Halstead M, Chanthavixay G, Trakooljul N, Wimmers K, Sahana G, Su GS, Lund MS, Fredholm M, Karlskov- Mortensen P, Ernst CW, Ross P, Tuggle CK, Fang LZ, Zhou HJ. Pig genome functional annotation enhances the biological interpretation of complex traits and human disease. Nat Commun, 2021, 12(1): 5848.
pmid: 34615879 |
| [32] |
González Morales N, Marescal O, Szikora S, Katzemich A, Correia-Mesquita T, Bíró P, Erdelyi M, Mihály J, Schöck F. The oxoglutarate dehydrogenase complex is involved in myofibril growth and Z-disc assembly in Drosophila. J Cell Sci, 2023, 136(13): jcs260717.
pmid: 37272588 |
| [33] |
Rahimi Y, Camporez JPG, Petersen MC, Pesta D, Perry RJ, Jurczak MJ, Cline GW, Shulman GI. Genetic activation of pyruvate dehydrogenase alters oxidative substrate selection to induce skeletal muscle insulin resistance. Proc Natl Acad Sci USA, 2014, 111(46): 16508-16513.
pmid: 25368185 |
| [34] |
Cairns G, Thumiah-Mootoo M, Abbasi MR, Gourlay M, Racine J, Larionov N, Prola A, Khacho M, Burelle Y. PINK1 deficiency alters muscle stem cell fate decision and muscle regenerative capacity. Stem Cell Reports, 2024, 19(5): 673-688.
pmid: 38579709 |
| [35] |
Li HZ, Yang WW, Cao WY, Yu ZK, Zhang GY, Long LZ, Guo H, Qu H, Fu CG, Chen KJ. Effects and mechanism of Kedaling tablets for atherosclerosis treatment based on network pharmacology, molecular docking and experimental study. J Ethnopharmacol, 2024, 319(Pt 1): 117108.
pmid: 37657772 |
| [36] | Huang Y. Genome-wide association analysis and genetic parameter estimation of carcass traits in Large White pigs[Dissertation]. Guangxi University, 2020. |
| 黄叶. 大白猪胴体性状全基因组关联分析及遗传参数估计[学位论文]. 广西大学, 2020. | |
| [37] |
Essén-Gustavsson B, Karlström K, Lundström K. Muscle fibre characteristics and metabolic response at slaughter in pigs of different halothane genotypes and their relation to meat quality. Meat Sci, 1992, 31(1): 1-11.
pmid: 22059505 |
| [38] |
Park J, Moon SS, Song SM, Cheng HL, Im C, Du LX, Kim GD. Comparative review of muscle fiber characteristics between porcine skeletal muscles. J Anim Sci Technol, 2024, 66(2): 251-265.
pmid: 38628685 |
| [39] |
Guo L, Han MM, Xu JF, Zhou WY, Shi HJ, Chen SS, Pang WJ, Zhang X, Duan YH, Yin YL, Li FN. snRNA-seq and spatial transcriptome reveal cell-cell crosstalk mediated metabolic regulation in porcine skeletal muscle. J Cachexia Sarcopenia Muscle, 2025, 16(2): e13752.
pmid: 40079370 |
| [40] |
Lee SH, Joo ST, Ryu YC. Skeletal muscle fiber type and myofibrillar proteins in relation to meat quality. Meat Sci, 2010, 86(1): 166-170.
pmid: 20605337 |
| [41] |
Li CC, Wang YN, Sun XH, Yang JJ, Ren YC, Jia JR, Yang GS, Liao MZ, Jin JJ, Shi XE. Identification of different myofiber types in pigs muscles and construction of regulatory networks. BMC Genomics, 2024, 25(1): 400.
pmid: 38658807 |
| [1] | 黄子莹, 李龙, 李倩倩, 刘向东, 李长春. lncRNA TCONS_00815878对猪骨骼肌卫星细胞分化的影响[J]. 遗传, 2019, 41(12): 1119-1128. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
