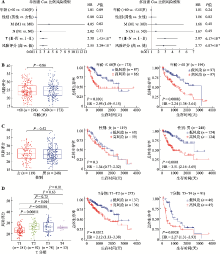
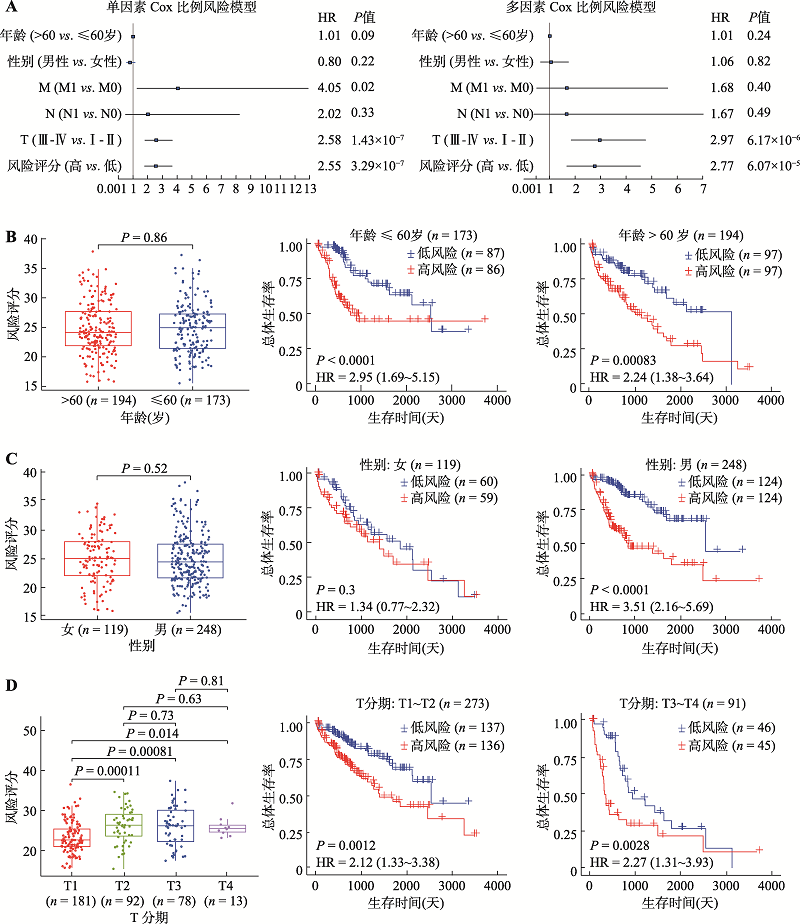
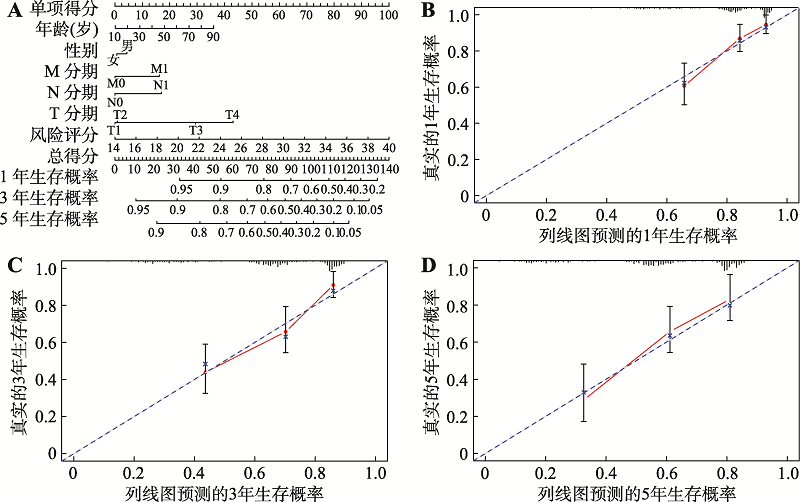
Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (2): 153-167.doi: 10.16288/j.yczz.21-416
• Research Article • Previous Articles Next Articles
Prognostic and predictive value of the hypoxia-associated long non-coding RNA signature in hepatocellular carcinoma
Min Cheng1,3( ), Jing Zhang4, Pengbo Cao2(
), Jing Zhang4, Pengbo Cao2( ), Gangqiao Zhou1,2,3(
), Gangqiao Zhou1,2,3( )
)
- 1. Department of Epidemiology, School of Public Health, Nanjing Medical University, Nanjing 211166, China
2. State Key Lab of Proteomics, National Center for Protein Sciences at Beijing, Institute of Radiation Medicine, Academy of Military Medical Sciences, Academy of Military Sciences, Beijing 100850, China
3. Center for Global Health, School of Public Health, Nanjing Medical University, Nanjing 211166, China
4. College of Life Science, Hebei University, Baoding 071002, China
-
Received:2021-12-02Revised:2022-01-11Online:2022-02-20Published:2022-01-19 -
Contact:Cao Pengbo,Zhou Gangqiao E-mail:18351990262@139.com;birchcpb@163.com;zhougq114@126.com -
Supported by:Supported by the National Key Research and Development Program of China No(2017YFA0504301);the Major Research Plan of the National Natural Science Foundation of China No(91440206)
Cite this article
Min Cheng, Jing Zhang, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of the hypoxia-associated long non-coding RNA signature in hepatocellular carcinoma[J]. Hereditas(Beijing), 2022, 44(2): 153-167.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Supplementary Table 1
Summary of clinical information of HCC patients in TCGA"
| 特征 | 样本量(n=367) | 比例(%) |
|---|---|---|
| 年龄(岁) | ||
| > 60 | 194 | 52.9 |
| ≤ 60 | 173 | 47.1 |
| 性别 | ||
| 女 | 119 | 32.4 |
| 男 | 248 | 67.6 |
| T分类 | ||
| T1(< 2厘米) | 181 | 49.3 |
| T2(2~5厘米) | 92 | 25.1 |
| T3(≥ 5厘米) | 78 | 21.3 |
| T4(靠近肝脏的血管和/或器官和/或内脏腹膜侵犯) | 13 | 3.5 |
| NA | 3 | 0.8 |
| N分类 | ||
| N0(附近的淋巴结没有癌细胞) | 249 | 67.8 |
| N1(肝脏附近淋巴结的癌细胞) | 4 | 1.1 |
| NA | 114 | 31.1 |
| M分类 | ||
| M0(没有癌细胞已经扩散到肝外的迹象) | 264 | 71.9 |
| M1(癌细胞存在于身体其他器官,如肺部或骨骼) | 3 | 0.8 |
| NA | 100 | 27.3 |
Supplementary Table 2
List of the hypoxia-related coding genes in tumors"
| MTX1 | ADORA2B | AK3 | ALDOA | ANGPTL4 | C20orf20 | MRPS17 | PGF | PGK1 |
|---|---|---|---|---|---|---|---|---|
| P4HA1 | PFKFB4 | PGAM1 | PVR | SLC16A1 | SLC2A1 | TEAD4 | TPBG | TPI1 |
| GAPD | GMFB | GSS | HES2 | HIG2 | IL8 | KCTD11 | KRT17 | PEDS1 |
| PSMA7 | PSMB7 | PSMD2 | PTGFRN | PYGL | RAN | RNF24 | RNPS1 | RUVBL2 |
| ANLN | VEGF | LOC56901 | S100A3 | B4GALT2 | VEZT | LRP2BP | SIP1 | BCAR1 |
| MGC14560 | SLC6A10 | BMS1L | ANKRD9 | MGC17624 | SLC6A8 | BNIP3 | C14orf156 | MGC2408 |
| HOMER1 | C15orf25 | MIF | SMILE | HSPC163 | CA12 | MRPL14 | SNX24 | IMP-2 |
| NUDT15 | SPTB | KIAA1393 | LDHA | LDLR | MGC2654 | MNAT1 | NDRG1 | NME1 |
| COL4A5 | CORO1C | CTEN | DKFZP564D166 | DPM2 | EIF2S1 | PAWR | PDZK11 | PLAU |
| PPARD | PPP2CZ | PPP4R1 | TFAP2C | TIMM23 | TMEM30B | TPD52L2 | VAPB | XPO5 |
| ANLN | BNC1 | C20orf20 | CA9 | CDKN3 | COL4A6 | DCBLD1 | ENO1 | FAM83B |
| GNAI1 | HIG2 | KCTD11 | KRT17 | LDHA | MPRS17 | P4HA1 | PGAM1 | PGK1 |
| AFARP1 | TUBB2 | LOC149464 | S100A10 | AD-003 | SLCO1B3 | CA9 | CDCA4 | PLEKHG3 |
| ALDOA | FOSL1 | SDC1 | SLC16A1 | SLC2A1 | TPI1 | VEGFA |
Supplementary Table 3
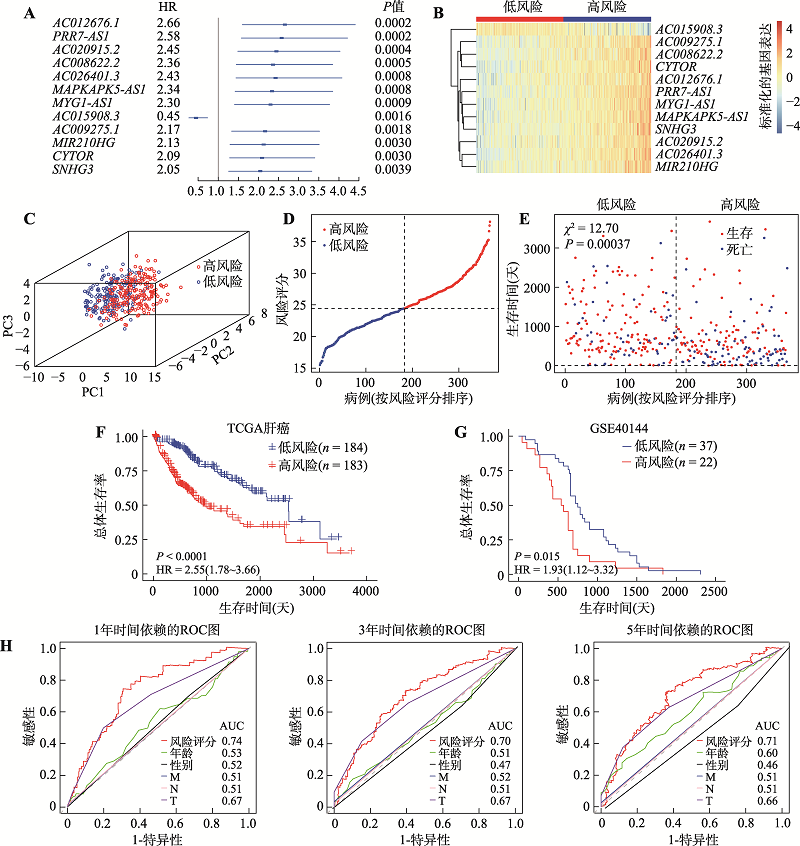
20 hypoxia-associated lncRNAs with prognostic value in HCCs"
| LncRNA | Rho | HR (95% CI) | P | 类型 |
|---|---|---|---|---|
| AC004816.1 | 0.6042 | 1.83 (1.29~2.60) | 0.00076 | 风险 |
| AC008622.2 | 0.8633 | 2.37 (1.66~3.40) | < 0.0001 | 风险 |
| AC009275.1 | 0.6005 | 1.82 (1.28~2.59) | 0.00083 | 风险 |
| AC012676.1 | 0.7518 | 2.12 (1.49~3.02) | < 0.0001 | 风险 |
| AC015908.3 | -0.7035 | 0.49 (0.35~0.71) | 0.0001 | 保护 |
| AC020915.2 | 0.7523 | 2.12 (1.48~3.04) | < 0.0001 | 风险 |
| AC026401.3 | 0.6128 | 1.85 (1.30~2.63) | 0.00068 | 风险 |
| AC073573.1 | -0.6068 | 0.55 (0.38~0.78) | 0.00093 | 保护 |
| AC114803.1 | 0.9514 | 2.59 (1.81~3.71) | < 0.0001 | 风险 |
| CYTOR | 0.7993 | 2.22 (1.56~3.18) | < 0.0001 | 风险 |
| DANCR | 0.7093 | 2.03 (1.43~2.90) | < 0.0001 | 风险 |
| GIHCG | 0.7331 | 2.08 (1.46~2.97) | < 0.0001 | 风险 |
| MAFG.DT | 0.5451 | 1.72 (1.22~2.44) | 0.0022 | 风险 |
| MAPKAPK5-AS1 | 0.9134 | 2.49 (1.73~2.59) | < 0.0001 | 风险 |
| MIR210HG | 0.7222 | 2.06 (1.44~2.95) | < 0.0001 | 风险 |
| MIR4435.2HG | 0.6980 | 2.01 (1.41~2.86) | 0.00011 | 风险 |
| MYG1-AS1 | 0.5395 | 1.72 (1.21~2.43) | 0.0025 | 风险 |
| PRR7-AS1 | 0.8126 | 2.25 (1.59~3.19) | < 0.0001 | 风险 |
| SNHG3 | 0.6218 | 1.86 (1.31~2.65) | 0.00054 | 风险 |
| TMEM220-AS1 | -0.4673 | 0.63 (0.44~0.89) | 0.0089 | 保护 |
Supplementary Table 4
GSEA results based on the risk score in HCCs from TCGA"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_RETROGRADE_VESICLE_MEDIATED_TRANSPORT_GOLGI_TO_ER | 77 | 2.34 | 0 |
| GO_NEGATIVE_REGULATION_OF_MITOTIC_CELL_CYCLE | 198 | 1.82 | 0 |
| GO_REGULATION_OF_PROTEASOMAL_UBIQUITIN_DEPENDENT_PROTEIN_CATABOLIC_ PROCESS | 146 | 1.81 | 0 |
| GO_POSITIVE_REGULATION_OF_CELL_CYCLE_PROCESS | 243 | 1.81 | 0 |
| GO_NUCLEAR_CHROMOSOME_SEGREGATION | 218 | 1.81 | 0 |
| GO_TUBULIN_BINDING | 263 | 1.78 | 0 |
| GO_MICROTUBULE_BASED_MOVEMENT | 199 | 1.77 | 0 |
| GO_MICROTUBULE | 397 | 1.77 | 0 |
| GO_SPINDLE_MIDZONE | 27 | 1.81 | 0.0020 |
| GO_REGULATION_OF_CELL_DIVISION | 266 | 1.80 | 0.0021 |
| GO_NEGATIVE_REGULATION_OF_ORGANELLE_ORGANIZATION | 384 | 1.77 | 0.0021 |
| GO_REGULATION_OF_DNA_DEPENDENT_DNA_REPLICATION | 41 | 1.81 | 0.0040 |
| GO_NUCLEAR_UBIQUITIN_LIGASE_COMPLEX | 42 | 1.81 | 0.0041 |
| GO_POSITIVE_REGULATION_OF_G1_S_TRANSITION_OF_MITOTIC_CELL_CYCLE | 24 | 1.78 | 0.0041 |
| GO_CONDENSED_CHROMOSOME | 183 | 1.77 | 0.0043 |
"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_RECOMBINATIONAL_REPAIR | 70 | 1.78 | 0.0063 |
| GO_CONDENSED_CHROMOSOME_CENTROMERIC_REGION | 94 | 1.78 | 0.0064 |
| GO_POSITIVE_REGULATION_OF_MITOTIC_NUCLEAR_DIVISION | 51 | 1.77 | 0.0064 |
| GO_SPINDLE_ASSEMBLY | 68 | 1.81 | 0.0081 |
| GO_HETEROCHROMATIN | 66 | 1.77 | 0.0083 |
| GO_CELL_CYCLE_G2_M_PHASE_TRANSITION | 132 | 1.81 | 0.0084 |
| GO_MITOTIC_CELL_CYCLE_CHECKPOINT | 138 | 1.80 | 0.013 |
| GO_SPINDLE_LOCALIZATION | 38 | 1.78 | 0.014 |
| GO_POSITIVE_REGULATION_OF_CHROMOSOME_SEGREGATION | 25 | 1.77 | 0.014 |
| GO_MEMBRANE_DISASSEMBLY | 46 | 1.79 | 0.021 |
| GO_REGULATION_OF_TELOMERE_MAINTENANCE | 62 | 1.81 | 0.023 |
| GO_CHROMOSOMAL_REGION | 310 | 1.79 | 0.023 |
| GO_SPLICEOSOMAL_COMPLEX | 163 | 1.78 | 0.033 |
| GO_RNA_SPLICING | 335 | 1.79 | 0.038 |
| GO_LIPID_OXIDATION | 68 | -2.15 | 0 |
| GO_MICROBODY | 132 | -2.13 | 0 |
| GO_FATTY_ACID_CATABOLIC_PROCESS | 71 | -2.13 | 0 |
| GO_FATTY_ACID_BETA_OXIDATION | 49 | -2.10 | 0 |
| GO_MICROBODY_PART | 92 | -2.08 | 0 |
| GO_COENZYME_BINDING | 175 | -2.07 | 0 |
| GO_ORGANIC_ACID_CATABOLIC_PROCESS | 202 | -2.03 | 0 |
| GO_FLAVIN_ADENINE_DINUCLEOTIDE_BINDING | 73 | -2.02 | 0 |
| GO_MICROBODY_MEMBRANE | 58 | -2.00 | 0 |
| GO_METHIONINE_METABOLIC_PROCESS | 18 | -1.99 | 0 |
| GO_COFACTOR_BINDING | 258 | -1.97 | 0 |
| GO_MICROBODY_LUMEN | 44 | -1.97 | 0 |
| GO_AMINO_ACID_BETAINE_METABOLIC_PROCESS | 18 | -1.94 | 0 |
| GO_LIPID_HOMEOSTASIS | 107 | -1.89 | 0 |
| GO_BILE_ACID_METABOLIC_PROCESS | 35 | -1.89 | 0 |
| GO_REGULATION_OF_FATTY_ACID_OXIDATION | 27 | -1.84 | 0 |
| GO_BILE_ACID_BIOSYNTHETIC_PROCESS | 20 | -1.82 | 0 |
| GO_CELLULAR_ALDEHYDE_METABOLIC_PROCESS | 83 | -1.82 | 0 |
| GO_ACYLGLYCEROL_HOMEOSTASIS | 29 | -1.82 | 0 |
| GO_MONOOXYGENASE_ACTIVITY | 91 | -1.80 | 0 |
| GO_DRUG_METABOLIC_PROCESS | 39 | -1.78 | 0 |
| GO_POSITIVE_REGULATION_OF_FATTY_ACID_METABOLIC_PROCESS | 33 | -1.78 | 0 |
| GO_NITROGEN_CYCLE_METABOLIC_PROCESS | 15 | -1.75 | 0 |
| GO_STEROID_HYDROXYLASE_ACTIVITY | 31 | -1.70 | 0 |
| GO_IRON_ION_BINDING | 158 | -1.70 | 0 |
| GO_EPOXYGENASE_P450_PATHWAY | 18 | -1.69 | 0 |
"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_OXYGEN_BINDING | 47 | -1.67 | 0 |
| GO_ARACHIDONIC_ACID_MONOOXYGENASE_ACTIVITY | 15 | -1.66 | 0 |
| GO_PEROXISOME_ORGANIZATION | 32 | -2.00 | 0.0020 |
| GO_PROTEIN_DEGLYCOSYLATION | 21 | -1.97 | 0.0020 |
| GO_REGULATION_OF_TRIGLYCERIDE_METABOLIC_PROCESS | 32 | -1.90 | 0.0020 |
| GO_SERINE_FAMILY_AMINO_ACID_METABOLIC_PROCESS | 41 | -1.89 | 0.0020 |
| GO_2_OXOGLUTARATE_METABOLIC_PROCESS | 20 | -1.88 | 0.0020 |
| GO_REGULATION_OF_TRIGLYCERIDE_BIOSYNTHETIC_PROCESS | 17 | -1.88 | 0.0020 |
| GO_CELLULAR_AMINO_ACID_CATABOLIC_PROCESS | 111 | -1.87 | 0.0020 |
| GO_ALPHA_AMINO_ACID_CATABOLIC_PROCESS | 94 | -1.87 | 0.0020 |
| GO_PYRIDOXAL_PHOSPHATE_BINDING | 51 | -1.86 | 0.0020 |
| GO_SULFUR_AMINO_ACID_METABOLIC_PROCESS | 40 | -1.84 | 0.0020 |
| GO_REGULATION_OF_CHOLESTEROL_METABOLIC_PROCESS | 22 | -1.84 | 0.0020 |
| GO_ACYL_COA_DEHYDROGENASE_ACTIVITY | 17 | -1.82 | 0.0020 |
| GO_GLYOXYLATE_METABOLIC_PROCESS | 27 | -1.81 | 0.0020 |
| GO_REGULATION_OF_MITOCHONDRIAL_FISSION | 17 | -1.73 | 0.0020 |
| GO_ENERGY_RESERVE_METABOLIC_PROCESS | 72 | -1.68 | 0.0020 |
| GO_BILE_ACID_TRANSMEMBRANE_TRANSPORTER_ACTIVITY | 15 | -1.66 | 0.0020 |
| GO_SULFUR_AMINO_ACID_BIOSYNTHETIC_PROCESS | 19 | -1.86 | 0.0021 |
| GO_REGULATION_OF_GLUCOSE_METABOLIC_PROCESS | 104 | -1.80 | 0.0021 |
| GO_BLOOD_COAGULATION_INTRINSIC_PATHWAY | 17 | -1.77 | 0.0021 |
| GO_ORGANIC_HYDROXY_COMPOUND_TRANSPORT | 155 | -1.67 | 0.0021 |
| GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_PAIRED_DONORS_WITH_INCORPORATION_OR_REDUCTION_OF_MOLECULAR_OXYGEN_REDUCED_FLAVIN_OR_FLAVOPROTEIN_AS_ONE_DONOR_AND_INCORPORATION_OF_ONE_ATOM_OF_OXYGEN | 26 | -1.65 | 0.0021 |
| GO_FATTY_ACYL_COA_BINDING | 30 | -1.87 | 0.0039 |
| GO_FATTY_ACID_BETA_OXIDATION_USING_ACYL_COA_DEHYDROGENASE | 18 | -1.79 | 0.0040 |
| GO_BILE_ACID_AND_BILE_SALT_TRANSPORT | 31 | -1.76 | 0.0040 |
| GO_GLUCAN_METABOLIC_PROCESS | 58 | -1.75 | 0.0040 |
| GO_REGULATION_OF_LIPID_CATABOLIC_PROCESS | 50 | -1.74 | 0.0040 |
| GO_CELLULAR_LIPID_CATABOLIC_PROCESS | 148 | -1.84 | 0.0041 |
| GO_SMALL_MOLECULE_CATABOLIC_PROCESS | 325 | -1.83 | 0.0041 |
| GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_PAIRED_DONORS_WITH_INCORPORATION_OR_REDUCTION_OF_MOLECULAR_OXYGEN | 149 | -1.75 | 0.0041 |
| GO_RESPONSE_TO_XENOBIOTIC_STIMULUS | 104 | -1.74 | 0.0041 |
| GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_PAIRED_DONORS_WITH_INCORPORATION_OR_REDUCTION_OF_MOLECULAR_OXYGEN_NAD_P_H_AS_ONE_DONOR_AND_INCORPORATION_OF_ONE_ATOM_OF_OXYGEN | 36 | -1.73 | 0.0041 |
| GO_BLOOD_COAGULATION_FIBRIN_CLOT_FORMATION | 24 | -1.73 | 0.0041 |
| GO_DRUG_TRANSMEMBRANE_TRANSPORT | 19 | -1.68 | 0.0041 |
| GO_GLUTAMATE_METABOLIC_PROCESS | 28 | -1.69 | 0.0042 |
| GO_ALPHA_AMINO_ACID_METABOLIC_PROCESS | 226 | -1.80 | 0.0060 |
| GO_PROTEIN_ACTIVATION_CASCADE | 67 | -1.78 | 0.0060 |
"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_ALDEHYDE_DEHYDROGENASE_NAD_ACTIVITY | 19 | -1.72 | 0.0060 |
| GO_ANDROGEN_METABOLIC_PROCESS | 30 | -1.72 | 0.0062 |
| GO_TRANSCRIPTION_FACTOR_ACTIVITY_DIRECT_LIGAND_REGULATED_SEQUENCE_ SPECIFIC_DNA_BINDING | 48 | -1.67 | 0.0062 |
| GO_BRANCHED_CHAIN_AMINO_ACID_METABOLIC_PROCESS | 23 | -1.78 | 0.0079 |
| GO_MANNOSIDASE_ACTIVITY | 15 | -1.76 | 0.0079 |
| GO_REACTIVE_NITROGEN_SPECIES_METABOLIC_PROCESS | 19 | -1.76 | 0.0079 |
| GO_STEROL_TRANSPORT | 50 | -1.69 | 0.0081 |
| GO_GLYCINE_METABOLIC_PROCESS | 17 | -1.68 | 0.0082 |
| GO_MONOCARBOXYLIC_ACID_TRANSMEMBRANE_TRANSPORTER_ACTIVITY | 45 | -1.64 | 0.0083 |
| GO_STEROID_METABOLIC_PROCESS | 232 | -1.78 | 0.0085 |
| GO_S_ADENOSYLMETHIONINE_METABOLIC_PROCESS | 18 | -1.73 | 0.010 |
| GO_CELLULAR_AMINO_ACID_BIOSYNTHETIC_PROCESS | 91 | -1.73 | 0.010 |
| GO_FATTY_ACID_METABOLIC_PROCESS | 287 | -1.70 | 0.010 |
| GO_TRANSMEMBRANE_RECEPTOR_PROTEIN_SERINE_THREONINE_KINASE_ACTIVITY | 17 | -1.68 | 0.010 |
| GO_BENZENE_CONTAINING_COMPOUND_METABOLIC_PROCESS | 24 | -1.68 | 0.010 |
| GO_REGULATION_OF_GLUCONEOGENESIS | 37 | -1.81 | 0.012 |
| GO_ALPHA_AMINO_ACID_BIOSYNTHETIC_PROCESS | 75 | -1.73 | 0.012 |
| GO_MONOCARBOXYLIC_ACID_METABOLIC_PROCESS | 491 | -1.71 | 0.012 |
| GO_THIOESTER_METABOLIC_PROCESS | 83 | -1.70 | 0.012 |
| GO_PROTEIN_LIPID_COMPLEX | 39 | -1.70 | 0.012 |
| GO_PLATELET_DENSE_GRANULE | 20 | -1.71 | 0.013 |
| GO_ARGININE_METABOLIC_PROCESS | 17 | -1.66 | 0.013 |
| GO_CELLULAR_AMINO_ACID_METABOLIC_PROCESS | 328 | -1.75 | 0.014 |
| GO_REGULATION_OF_CELLULAR_KETONE_METABOLIC_PROCESS | 170 | -1.68 | 0.014 |
| GO_TRICARBOXYLIC_ACID_METABOLIC_PROCESS | 37 | -1.82 | 0.016 |
| GO_ASPARTATE_FAMILY_AMINO_ACID_METABOLIC_PROCESS | 55 | -1.80 | 0.016 |
| GO_REGULATION_OF_PROTEIN_ACTIVATION_CASCADE | 34 | -1.76 | 0.016 |
| GO_NEGATIVE_REGULATION_OF_MITOCHONDRION_ORGANIZATION | 39 | -1.71 | 0.016 |
| GO_POSITIVE_REGULATION_OF_TRIGLYCERIDE_METABOLIC_PROCESS | 20 | -1.71 | 0.017 |
| GO_RESPONSE_TO_MERCURY_ION | 15 | -1.69 | 0.017 |
| GO_REGULATION_OF_FATTY_ACID_METABOLIC_PROCESS | 85 | -1.65 | 0.017 |
| GO_STEROL_HOMEOSTASIS | 57 | -1.77 | 0.019 |
| GO_STEROL_METABOLIC_PROCESS | 121 | -1.71 | 0.019 |
| GO_REGULATION_OF_LIPOPROTEIN_LIPASE_ACTIVITY | 15 | -1.64 | 0.019 |
| GO_MITOCHONDRIAL_MATRIX | 406 | -1.84 | 0.020 |
| GO_DICARBOXYLIC_ACID_METABOLIC_PROCESS | 99 | -1.73 | 0.020 |
| GO_PROTEIN_HOMOTETRAMERIZATION | 59 | -1.73 | 0.020 |
| GO_COMPLEMENT_ACTIVATION | 45 | -1.67 | 0.020 |
| GO_POSITIVE_REGULATION_OF_LIPID_CATABOLIC_PROCESS | 25 | -1.69 | 0.021 |
| GO_COFACTOR_METABOLIC_PROCESS | 329 | -1.66 | 0.024 |
"
| 基因集 | 基因集大小 | NES | P |
|---|---|---|---|
| GO_OXIDOREDUCTASE_ACTIVITY_ACTING_ON_THE_CH_CH_GROUP_OF_DONORS | 57 | -1.77 | 0.025 |
| GO_QUATERNARY_AMMONIUM_GROUP_TRANSPORT | 18 | -1.74 | 0.025 |
| GO_SIGNAL_PEPTIDE_PROCESSING | 24 | -1.71 | 0.027 |
| GO_GLUCAN_BIOSYNTHETIC_PROCESS | 25 | -1.68 | 0.027 |
| GO_RETROGRADE_TRANSPORT_VESICLE_RECYCLING_WITHIN_GOLGI | 23 | -1.72 | 0.028 |
| GO_NAD_BINDING | 53 | -1.69 | 0.029 |
| GO_NUCLEOSIDE_BISPHOSPHATE_METABOLIC_PROCESS | 37 | -1.70 | 0.030 |
| GO_ENDOCYTIC_VESICLE_LUMEN | 17 | -1.64 | 0.030 |
| GO_LIGASE_ACTIVITY_FORMING_CARBON_SULFUR_BONDS | 40 | -1.65 | 0.035 |
| GO_TRIGLYCERIDE_RICH_LIPOPROTEIN_PARTICLE | 19 | -1.65 | 0.035 |
| GO_ASPARTATE_FAMILY_AMINO_ACID_BIOSYNTHETIC_PROCESS | 23 | -1.66 | 0.036 |
| GO_COENZYME_A_METABOLIC_PROCESS | 17 | -1.65 | 0.041 |
| GO_2_IRON_2_SULFUR_CLUSTER_BINDING | 21 | -1.65 | 0.044 |
Supplementary Table 5
Primers for RT-qPCR assays"
| 基因 | 正义(5′→3′) | 反义 (5′→3′) |
|---|---|---|
| MIR210HG | TGAGTAGGAACTCTGGGCGA | CCACAATGGGAAGGAGGCAT |
| HIF1A | AGAGGTTGAGGGACGGAGAT | GCACCAAGCAGGTCATAGGT |
| TGFB | GTCTCCCAAGGAAAGGTAGG | CTCTTGAGTCCCTCGCATCC |
| AKT | GCGGCAGGACCGAGC | AGGTCTTGATGTACTCCCCTCG |
| VEGFA | GTCCTGGAGCGTGTACGTTG | CTTCCGGGCTCGGTGATTTA |
| ACTIN | AGAGCCTCGCCTTTGCCGAT | AGAGCCTCGCCTTTGCCGAT |
| [1] |
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin, 2015, 65(2): 87-108.S
doi: 10.3322/caac.21262 |
| [2] |
Yuen VW, Wong CC. Hypoxia-inducible factors and innate immunity in liver cancer. J Clin Invest, 2020, 130(10):5052-5062.
doi: 10.1172/JCI137553 |
| [3] |
Shimamoto K, Tanimoto K, Fukazawa T, Nakamura H, Kanai A, Bono H, Ono H, Eguchi H, Hirohashi N. GLIS1, a novel hypoxia-inducible transcription factor, promotes breast cancer cell motility via activation of WNT5A. Carcinogenesis, 2020, 41(9):1184-1194.
doi: 10.1093/carcin/bgaa010 pmid: 32047936 |
| [4] |
Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis, 2014, 35(3):507-514.
doi: 10.1093/carcin/bgt405 |
| [5] |
Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA, 2014, 20(12):1844-1849.
doi: 10.1261/rna.047332.114 pmid: 25316907 |
| [6] |
Wang Y, Chen WY, Lian JY, Zhang HB, Yu B, Zhang MJ, Wei FQ, Wu JH, Jiang JX, Jia YS, Mo F, Zhang SR, Liang XD, Mou XZ, Tang JM. The lncRNA PVT1 regulates nasopharyngeal carcinoma cell proliferation via activating the KAT2A acetyltransferase and stabilizing HIF-1α. Cell Death Differ, 2020, 27(2):695-710.
doi: 10.1038/s41418-019-0381-y |
| [7] |
Wang XW, Li L, Zhao KM, Lin QY, Li HY, Xue XT, Ge WJ, He HJ, Liu D, Xie H, Wu Q, Hu Y. A novel LncRNA HITT forms a regulatory loop with HIF-1α to modulate angiogenesis and tumor growth. Cell Death Differ, 2020, 27(4):1431-1446.
doi: 10.1038/s41418-019-0449-8 |
| [8] |
Zeng Z, Xu FY, Zheng H, Cheng P, Chen QY, Ye Z, Zhong JX, Deng SJ, Liu ML, Huang K, Li Q, Li W, Hu YH, Wang F, Wang CY, Zhao G. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1α. Theranostics, 2019, 9(18):5298-5314.
doi: 10.7150/thno.34559 pmid: 31410216 |
| [9] |
Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z, He C, Liu ML, Huang K, Zhong JX, Xu FY, Li Q, Liu Y, Wang CY, Zhao G. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene, 2018, 37(44):5811-5828.
doi: 10.1038/s41388-018-0382-1 |
| [10] |
Liang YR, Song XJ, Li YM, Chen B, Zhao WJ, Wang LJ, Zhang HW, Liu Y, Han DW, Zhang N, Ma TT, Wang YJ, Ye FZ, Luo D, Li XY, Yang QF. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer, 2020, 19(1):85.
doi: 10.1186/s12943-020-01206-5 |
| [11] |
Dong L, Cao X, Luo Y, Zhang GQ, Zhang DD. A positive feedback loop of lncRNA DSCR8/miR-98-5p/STAT3/HIF- 1α plays a role in the progression of ovarian cancer. Front Oncol, 2020, 10:1713.
doi: 10.3389/fonc.2020.01713 |
| [12] |
Hua Q, Mi BM, Xu F, Wen J, Zhao L, Liu JJ, Huang G. Hypoxia-induced lncRNA-AC020978 promotes proliferation and glycolytic metabolism of non-small cell lung cancer by regulating PKM2/HIF-1α axis. Theranostics, 2020, 10(11):4762-4778.
doi: 10.7150/thno.43839 |
| [13] |
Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, Shah KA, Cox GJ, Corbridge RJ, Homer JJ, Musgrove B, Slevin N, Sloan P, Price P, West CM, Harris AL. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res, 2007, 67(7):3441-3449.
doi: 10.1158/0008-5472.CAN-06-3322 |
| [14] |
Eustace A, Mani N, Span PN, Irlam JJ, Taylor J, Betts GNJ, Denley H, Miller CJ, Homer JJ, Rojas AM, Hoskin PJ, Buffa FM, Harris AL, Kaanders JHAM, West CML. A 26-gene hypoxia signature predicts benefit from hypoxia- modifying therapy in laryngeal cancer but not bladder cancer. Clin Cancer Res, 2013, 19(17):4879-4888.
doi: 10.1158/1078-0432.CCR-13-0542 pmid: 23820108 |
| [15] |
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics, 2013, 14:7.
doi: 10.1186/1471-2105-14-7 pmid: 23323831 |
| [16] | Sun ZL, Jing CY, Xiao CT, Li TC. An autophagy-related long non-coding RNA prognostic signature accurately predicts survival outcomes in bladder urothelial carcinoma patients. Aging (Albany NY), 2020, 12(15):15624-15637. |
| [17] |
Steyerberg EW, Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J, 2014, 35(29):1925-1931.
doi: 10.1093/eurheartj/ehu207 pmid: 24898551 |
| [18] |
Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med, 2013, 32(30):5381-5397.
doi: 10.1002/sim.5958 pmid: 24027076 |
| [19] |
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA, 2005, 102(43):15545-15550.
doi: 10.1073/pnas.0506580102 |
| [20] |
Zhang CJ, Zheng YX, Li X, Hu X, Qi F, Luo J. Genome-wide mutation profiling and related risk signature for prognosis of papillary renal cell carcinoma. Ann Transl Med, 2019, 7(18):427.
doi: 10.21037/atm |
| [21] |
Tang FZ, Li CW, Kang BX, Gao G, Li C, Zhang ZM. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res, 2017, 45(W1):W98-W102.
doi: 10.1093/nar/gkx247 |
| [22] |
Kim S, Kang D, Huo ZG, Park Y, Tseng GC. Meta- analytic principal component analysis in integrative omics application. Bioinformatics, 2018, 34(8):1321-1328.
doi: 10.1093/bioinformatics/btx765 |
| [23] | Tang R, Wu JC, Zheng LM, Li ZR, Zhou KL, Zhang ZS, Xu DF, Chen C. Long noncoding RNA RUSC1-AS-N indicates poor prognosis and increases cell viability in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci, 2018, 22(2):388-396. |
| [24] | Torgovnick A, Schumacher B. DNA repair mechanisms in cancer development and therapy. Front Genet, 2015, 6:157. |
| [25] |
Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F. microRNA: emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther, 2010, 125(1):92-104.
doi: 10.1016/j.pharmthera.2009.10.003 |
| [26] |
Feng S, He AB, Wang DL, Kang B. Diagnostic significance of miR-210 as a potential tumor biomarker of human cancer detection: an updated pooled analysis of 30 articles. Onco Targets Ther, 2019, 12:479-493.
doi: 10.2147/OTT |
| [27] |
Kim JH, Park SG, Song SY, Kim JK, Sung JH. Reactive oxygen species-responsive miR-210 regulates proliferation and migration of adipose-derived stem cells via PTPN2. Cell Death Dis, 2013, 4(4):e588.
doi: 10.1038/cddis.2013.117 |
| [28] |
Yang W, Sun T, Cao JP, Liu FJ, Tian Y, Zhu W. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res, 2012, 318(8):944-954.
doi: 10.1016/j.yexcr.2012.02.010 pmid: 22387901 |
| [29] |
Ying Q, Liang LH, Guo WJ, Zha RP, Tian Q, Huang SL, Yao J, Ding J, Bao MY, Ge C, Yao M, Li JJ, He XH. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology, 2011, 54(6):2064-2075.
doi: 10.1002/hep.24614 |
| [30] |
Wu H, Wang T, Liu YQ, Li X, Xu SL, Wu CT, Zou HB, Cao MF, Jin GX, Lang JY, Wang B, Liu BH, Luo XL, Xu C. Mitophagy promotes sorafenib resistance through hypoxia-inducible ATAD3A dependent axis. J Exp Clin Cancer Res, 2020, 39(1):274.
doi: 10.1186/s13046-020-01768-8 |
| [31] |
Ebright RY, Zachariah MA, Micalizzi DS, Wittner BS, Niederhoffer KL, Nieman LT, Chirn B, Wiley DF, Wesley B, Shaw B, Nieblas-Bedolla E, Atlas L, Szabolcs A, Iafrate AJ, Toner M, Ting DT, Brastianos PK, Haber DA, Maheswaran S. HIF1A signaling selectively supports proliferation of breast cancer in the brain. Nat Commun, 2020, 11(1):6311.
doi: 10.1038/s41467-020-20144-w |
| [32] |
Lin YT, Wu KJ. Epigenetic regulation of epithelial- mesenchymal transition: focusing on hypoxia and TGF-β signaling. J Biomed Sci, 2020, 27(1):39.
doi: 10.1186/s12929-020-00632-3 |
| [33] |
Wang XX, Che XF, Yu Y, Cheng Y, Bai M, Yang ZC, Guo QQ, Xie XC, Li DN, Guo M, Hou KZ, Guo WD, Qu XJ, Cao L. Hypoxia-autophagy axis induces VEGFA by peritoneal mesothelial cells to promote gastric cancer peritoneal metastasis through an integrin α5-fibronectin pathway. J Exp Clin Cancer Res, 2020, 39(1):221.
doi: 10.1186/s13046-020-01703-x |
| [34] |
Ramapriyan R, Caetano MS, Barsoumian HB, Mafra ACP, Zambalde EP, Menon H, Tsouko E, Welsh JW, Cortez MA. Altered cancer metabolism in mechanisms of immunotherapy resistance. Pharmacol Ther, 2019, 195:162-171.
doi: 10.1016/j.pharmthera.2018.11.004 |
| [35] |
Chen BW, Zhou Y, Wei T, Wen L, Zhang YB, Shen SC, Zhang J, Ma T, Chen W, Ni L, Wang Y, Bai XL, Liang TB. lncRNA-POIR promotes epithelial-mesenchymal transition and suppresses sorafenib sensitivity simultaneously in hepatocellular carcinoma by sponging miR-182-5p. J Cell Biochem, 2021, 122(1):130-142.
doi: 10.1002/jcb.v122.1 |
| [36] |
Sun Z, Xue SL, Zhang MY, Xu H, Hu XM, Chen SH, Liu YY, Guo MZ, Cui HM. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene, 2020, 39(45):6906-6919.
doi: 10.1038/s41388-020-01475-w |
| [37] |
Cai HY, Zhang Y, Zhang HY, Cui C, Li CH, Lu SC. Prognostic role of tumor mutation burden in hepatocellular carcinoma after radical hepatectomy. J Surg Oncol, 2020, 121(6):1007-1014.
doi: 10.1002/jso.v121.6 |
| [38] |
Maleki Vareki S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer, 2018, 6(1):157.
doi: 10.1186/s40425-018-0479-7 pmid: 30587233 |
| [39] |
Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of a six-gene signature predicting overall survival for hepatocellular carcinoma. Cancer Cell Int, 2019, 19:138.
doi: 10.1186/s12935-019-0858-2 |
| [40] |
Li XY, Jin F, Li Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J Cell Mol Med, 2021, 25(1):4-14.
doi: 10.1111/jcmm.v25.1 |
| [41] |
Li XY, Li Y, Yu XM, Jin F. Identification and validation of stemness-related lncRNA prognostic signature for breast cancer. J Transl Med, 2020, 18(1):331.
doi: 10.1186/s12967-020-02497-4 |
| [42] |
Jin LP, Li CY, Liu T, Wang L. A potential prognostic prediction model of colon adenocarcinoma with recurrence based on prognostic lncRNA signatures. Hum Genomics, 2020, 14(1):24.
doi: 10.1186/s40246-020-00270-8 |
| [43] |
Niklasson CU, Fredlund E, Monni E, Lindvall JM, Kokaia Z, Hammarlund EU, Bronner ME, Mohlin S. Hypoxia inducible factor-2α importance for migration, proliferation, and self-renewal of trunk neural crest cells. Dev Dyn, 2021, 250(2):191-236.
doi: 10.1002/dvdy.v250.2 |
| [44] |
Liu OH, Kiema M, Beter M, Ylä-Herttuala S, Laakkonen JP, Kaikkonen MU. Hypoxia-mediated regulation of histone demethylases affects angiogenesis-associated functions in endothelial cells. Arterioscler Thromb Vasc Biol, 2020, 40(11):2665-2677.
doi: 10.1161/ATVBAHA.120.315214 |
| [45] |
Renfrow JJ, Soike MH, West JL, Ramkissoon SH, Metheny-Barlow L, Mott RT, Kittel CA, D'agostino Jr RB, Tatter SB, Laxton AW, Frenkel MB, Hawkins GA, Herpai D, Sanders S, Sarkaria JN, Lesser GJ, Debinski W, Strowd RE. Attenuating hypoxia driven malignant behavior in glioblastoma with a novel hypoxia-inducible factor 2 alpha inhibitor. Sci Rep, 2020, 10(1):15195.
doi: 10.1038/s41598-020-72290-2 pmid: 32938997 |
| [46] |
Yu AJ, Zhao LW, Kang QM, Li J, Chen K, Fu H. Transcription factor HIF1α promotes proliferation, migration, and invasion of cholangiocarcinoma via long noncoding RNA H19/microRNA-612/Bcl-2 axis. Transl Res, 2020, 224:26-39.
doi: 10.1016/j.trsl.2020.05.010 |
| [47] |
Wang L, Sun LK, Liu RY, Mo HY, Niu YS, Chen TX, Wang YF, Han SS, Tu KS, Liu QG. Long non-coding RNA MAPKAPK5-AS1/PLAGL2/HIF-1α signaling loop promotes hepatocellular carcinoma progression. J Exp Clin Cancer Res, 2021, 40(1):72.
doi: 10.1186/s13046-021-01868-z |
| [48] | Hu B, Yang XB, Yang X, Sang XT. LncRNA CYTOR affects the proliferation, cell cycle and apoptosis of hepatocellular carcinoma cells by regulating the miR-125b-5p/KIAA1522 axis. Aging (Albany NY), 2020, 13(2):2626-2639. |
| [49] |
Wei F, Wang Y, Zhou Y, Li Y. Long noncoding RNA CYTOR triggers gastric cancer progression by targeting miR-103/RAB10. Acta Biochim Biophys Sin (Shanghai), 2021, 53(8):1044-1054.
doi: 10.1093/abbs/gmab071 |
| [50] |
Yang JL, Ma Q, Zhang MM, Zhang WF. LncRNA CYTOR drives L-OHP resistance and facilitates the epithelial- mesenchymal transition of colon carcinoma cells via modulating miR-378a-5p/SERPINE1. Cell Cycle, 2021, 20(14):1415-1430.
doi: 10.1080/15384101.2021.1934626 |
| [51] |
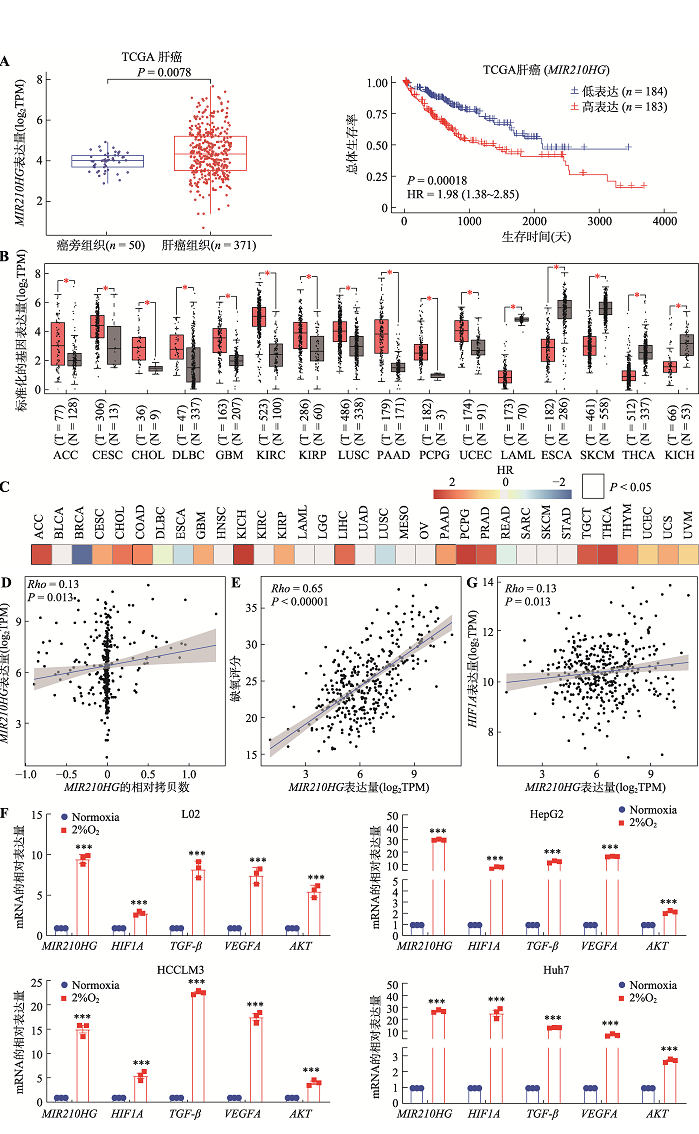
Wang Y, Li WJ, Chen XY, Li Y, Wen PH, Xu F. MIR210HG predicts poor prognosis and functions as an oncogenic lncRNA in hepatocellular carcinoma. Biomed Pharmacother, 2019, 111:1297-1301.
doi: S0753-3322(18)37945-9 pmid: 30841443 |
| [52] |
He ZY, Dang J, Song AL, Cui X, Ma ZJ, Zhang ZT. Identification of LINC01234 and MIR210HG as novel prognostic signature for colorectal adenocarcinoma. J Cell Physiol, 2019, 234(5):6769-6777.
doi: 10.1002/jcp.v234.5 |
| [53] |
Ma J, Kong FF, Yang D, Yang H, Wang CC, Cong R, Ma XX. LncRNA MIR210HG promotes the progression of endometrial cancer by sponging miR-337-3p/137 via the HMGA2-TGF-β/Wnt pathway. Mol Ther Nucleic Acids, 2021, 24:905-922.
doi: 10.1016/j.omtn.2021.04.011 |
| [54] | Bu L, Zhang LB, Tian M, Zheng ZB, Tang HJ, Yang QJ. LncRNA MIR210HG facilitates non-small cell lung cancer progression through directly regulation of miR-874/STAT3 axis. Dose Response, 2020, 18(3):1559325820918052. |
| [55] | Wang AH, Jin CH, Cui GY, Li HY, Wang Y, Yu JJ, Wang RF, Tian XY. MIR210HG promotes cell proliferation and invasion by regulating miR-503-5p/TRAF4 axis in cervical cancer. Aging (Albany NY), 2020, 12(4):3205-3217. |
| [56] |
Yu TZ, Li GP, Wang CG, Gong GQ, Wang LW, Li CY, Chen Y, Wang XL. MIR210HG regulates glycolysis, cell proliferation, and metastasis of pancreatic cancer cells through miR-125b-5p/HK2/PKM2 axis. RNA Biol, 2021, 18(12):2513-2530.
doi: 10.1080/15476286.2021.1930755 |
| [57] |
Du Y, Wei N, Ma RL, Jiang SH, Song D. Long noncoding RNA MIR210HG promotes the warburg effect and tumor growth by enhancing HIF-1α translation in triple-negative breast cancer. Front Oncol, 2020, 10:580176.
doi: 10.3389/fonc.2020.580176 |
| [1] | Changgui Lei, Xueyuan Jia, Wenjing Sun. Establish six-gene prognostic model for glioblastoma based on multi-omics data of TCGA database [J]. Hereditas(Beijing), 2021, 43(7): 665-679. |
| [2] | Hongbo Luo, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of a DNA methylation-driven transcriptional signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2020, 42(8): 775-787. |
| [3] | Huawei Zhang, Xingyu Meng, anfeng Li, Yuying Yang, Huaji Qiu. Long non-coding RNAs: Emerging regulators of antiviral innate immune responses [J]. Hereditas(Beijing), 2018, 40(7): 525-533. |
| [4] | Junxia Zou, Ke Chen. Roles and molecular mechanisms of hypoxia-inducible factors in renal cell carcinoma [J]. Hereditas(Beijing), 2018, 40(5): 341-356. |
| [5] | Rui Zhou,Yixin Wang,Keren Long,Anan Jiang,Long Jin. Regulatory mechanism for lncRNAs in skeletal muscle development and progress on its research in domestic animals [J]. Hereditas(Beijing), 2018, 40(4): 292-304. |
| [6] | Chang Lu, Yinhua Huang. Progress in long non-coding RNAs in animals [J]. Hereditas(Beijing), 2017, 39(11): 1054-1065. |
| [7] | Fang Yang, Baoyun Zhang, Guangde Feng, Wei Xiang, Yunxia Ma, Hang Chen, Mingxing Chu, Pingqing Wang. A mechanistic review of how hypoxic mircroenvironment regulates mammalian ovulation [J]. HEREDITAS(Beijing), 2016, 38(2): 109-117. |
| [8] | Jingqiu Li, Jie Yang, Ping Zhou, Yanping Le, Zhaohui Gong. The biological functions and regulations of competing endogenous RNA [J]. HEREDITAS(Beijing), 2015, 37(8): 756-764. |
| [9] | Lingyun Sun, Xingyu Li, Zhiwei Sun. Progress of epigenetics and its therapeutic application in hepatocellular carcinoma [J]. HEREDITAS(Beijing), 2015, 37(6): 517-527. |
| [10] | Xiaoqing Huang,Dandan Li,Juan Wu. Long non-coding RNAs in plants [J]. HEREDITAS(Beijing), 2015, 37(4): 344-359. |
| [11] | Feng Yang, Fan Yi, Huiqing Cao, Zicai Liang, Quan Du. The emerging landscape of long non-coding RNAs [J]. HEREDITAS(Beijing), 2014, 36(5): 456-468. |
| [12] | Ling Li, Xu Song. In vivo functions of long non-coding RNAs [J]. HEREDITAS, 2014, 36(3): 228-236. |
| [13] | LI Qian, LIU Shu-Yuan, LIN Ke-Qin, SUN Hao, YU Liang, HUANG Xiao-Qin, CHU Jia-You, YANG Zhao-Qing. Association between six single nucleotide polymorphisms of EGLN1 gene and adaptation to high-altitude hypoxia [J]. HEREDITAS, 2013, 35(8): 992-998. |
| [14] | XIA Tian, XIAO Bing-Xiu, GUO Jun-Ming. Acting mechanisms and research methods of long noncoding RNAs [J]. HEREDITAS, 2013, 35(3): 269-280. |
| [15] | ZHANG Hao, CHAMBA Yangzom, ZHAO Chun-Jiang, BAO Hai-Gang, LING Yao, WU Chang-Xin. Function of inducible nitric oxide synthase on adaptability to hypoxia in Tibetan chicken [J]. HEREDITAS, 2009, 31(4): 400-406. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||