Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (12): 1143-1155.doi: 10.16288/j.yczz.20-178
• Review • Previous Articles Next Articles
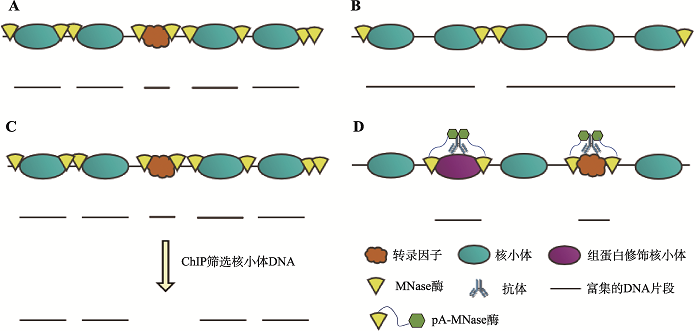
Resolving nucleosomal positioning and occupancy with MNase-seq
- Bio-ID Center, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai 200240, China
-
Received:2020-09-04Revised:2020-10-18Online:2020-12-17Published:2020-11-03 -
Contact:Li Xinhui E-mail:xhli@sjtu.edu.cn -
Supported by:Supported by the National Natural Science Foundation of China No(81972909)
Cite this article
Weihang Deng, Xinhui Li. Resolving nucleosomal positioning and occupancy with MNase-seq[J]. Hereditas(Beijing), 2020, 42(12): 1143-1155.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Technology for the study of nucleosome and chromatin structures"
| 技术名称 | 测序类型 | 实验流程 | 优势 | 缺陷 | 参考文献 |
|---|---|---|---|---|---|
| MNase-seq | 双端或 单端测序 | 1. 对染色质进行MNase酶切 2. 文库构建 3. 凝胶电泳筛选单核小体长度片段 | 1. 可以对全基因组范围内核小体进行测量 2. 技术难度较小 3. 酶切特性使其具有较高分辨率 | 1. 传统方法需要大量 细胞 2. 无法分辨核小体与其他DNA-蛋白质复合物 3. 容易造成技术误差 | [7,22~25] |
| ChIP-seq | 双端或 单端测序 | 1. 甲醛交联 2. 超声将染色质片段化 3. 利用特异性抗体沉淀特定蛋白质 | 1. 方法较成熟,适用范围 广泛 2. 技术难度较低 3. 针对特定的蛋白靶点, 特异性较高 | 1. 分辨率较低 2. 数据质量依赖抗体质量,筛选抗体费时费力 3. 成本较高 | [11,26,27] |
| ATAC-seq | 双端测序 | 1. 利用Tn5转座酶将开放染色质区域的DNA片段化并加上接头 2. 文库构建并上机测序 | 1. 灵敏性高,低细胞起始量 2. 方法简单,耗时短 3. 分辨率较高 4. 重复性较好 | 1. Tn5酶价格昂贵 2. 常规数据分析方法不可用或存在限制 | [28~30] |
| DNase-seq | 双端或 单端测序 | 1. 利用DNaseⅠ切割开放染色质区域 2. 连接接头、片段筛选 | 1. 分辨率较高 2. 实验方法较简单 | 1. 样品准备复杂,实验流程耗时 2. 需要大量细胞 3. 需精确控制酶量 | [31,32] |
| NoMe-seq | 双端测序 | 1. 利用GpC甲基转移酶处理固定的染色质 2. DNA片段的重亚硫酸盐转化 3. 文库构建并上机测序 | 可对同一样品同时进行核小体定位及DNA甲基化程度的分析 | 1. 需要大量细胞 2. 数据分析存在挑战 | [33,34] |
Table 1
| [1] | Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ . Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature, 1997,389(6648):251-260. |
| [2] | Zhou KD, Gaullier G, Luger K . Nucleosome structure and dynamics are coming of age. Nat Struct Mol Biol, 2019,26(1):3-13. |
| [3] | Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, Moore IK, Wang JZ, Widom J . A genomic code for nucleosome positioning. Nature, 2006,442(7104):772-778. |
| [4] | Noll M . Subunit structure of chromatin. Nature, 1974,251(5472):249-251. |
| [5] | Lohr D, Kovacic RT, Van Holde KE . Quantitative analysis of the digestion of yeast chromatin by staphylococcal nuclease. Biochemistry, 1977,16(3):463-471. |
| [6] | Fan XC, Moqtaderi Z, Jin Y, Zhang Y, Liu XS, Struhl K . Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3’-end formation. Proc Natl Acad Sci USA, 2010,107(42):17945-17950. |
| [7] | Tsompana M, Buck MJ . Chromatin accessibility: a window into the genome. Epigenetics Chromatin, 2014,7(1):33. |
| [8] | Zhang WL, Jiang JM . Application of MNase-Seq in the global mapping of nucleosome positioning in plants. Methods Mol Biol, 2018,1830:353-366. |
| [9] | Pajoro A, Muiño JM, Angenent GC, Kaufmann K . Profiling nucleosome occupancy by MNase-seq: experimental protocol and computational analysis. Methods Mol Biol, 2018,1675:167-181. |
| [10] | Gaffney DJ , McVicker G, Pai AA, Fondufe-Mittendorf YN, Lewellen N, Michelini K, Widom J, Gilad Y, Pitchard JK. Controls of nucleosome positioning in the human genome. PLoS Genet, 2012,8(11):e1003036. |
| [11] | Zhang ZH, Pugh BF . High-resolution genome-wide mapping of the primary structure of chromatin. Cell, 2011,144(2):175-186. |
| [12] | Klein DC, Hainer SJ . Genomic methods in profiling DNA accessibility and factor localization. Chromosome Res, 2020,28(1):69-85. |
| [13] | Chen WZ, Liu Y, Zhu SS, Green CD, Wei G, Han JDJ . Improved nucleosome-positioning algorithm iNPS for accurate nucleosome positioning from sequencing data. Nat Commun, 2014,5:4909. |
| [14] | Fu K, Tang QZ, Feng JX, Liu XS, Zhang Y . DiNuP: a systematic approach to identify regions of differential nucleosome positioning. Bioinformatics, 2012,28(15):1965-1971. |
| [15] | Yuan GC, Liu JS . Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput Biol, 2008,4(1):e13. |
| [16] | Wal M, Pugh BF . Genome-wide mapping of nucleosome positions in yeast using high-resolution MNase ChIP-Seq. Methods Enzymol, 2012,513:233-250. |
| [17] | Ocampo J, Cui F, Zhurkin VB, Clark DJ . The proto-chromatosome: A fundamental subunit of chromatin? Nucleus, 2016,7(4):382-387. |
| [18] | Mieczkowski J, Cook A, Bowman SK, Mueller B, Alver BH, Kundu S, Deaton AM, Urban JA, Larschan E, Park PJ, Kingston RE, Tolstorukov MY . MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat Commun, 2016,7:11485. |
| [19] | Zhao HN, Zhang WL, Zhang T, Lin Y, Hu YD, Fang C, Jiang JM . Genome-wide MNase hypersensitivity assay unveils distinct classes of open chromatin associated with H3K27me3 and DNA methylation in Arabidopsis thaliana. Genome Biol, 2020,21(1):24. |
| [20] | Baldi S, Krebs S, Blum H, Becker PB . Genome-wide measurement of local nucleosome array regularity and spacing by nanopore sequencing. Nat Struct Mol Biol, 2018,25(9):894-901. |
| [21] | Skene PJ, Henikoff S . An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife, 2017,6:e21856. |
| [22] | Rizzo JM, Bard JE, Buck MJ . Standardized collection of MNase-seq experiments enables unbiased dataset comparisons. BMC Mol Biol, 2012,13:15. |
| [23] | Rizzo JM, Sinha S . Analyzing the global chromatin structure of keratinocytes by MNase-seq. Methods Mol Biol, 2014,1195:49-59. |
| [24] | Cui KR, Zhao KJ . Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol Biol, 2012,833:413-419. |
| [25] | Meyer CA, Liu XS . Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet, 2014,15(11):709-721. |
| [26] | Park PJ . ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet, 2009,10(10):669-680. |
| [27] | Mardis ER . ChIP-seq: welcome to the new frontier. Nat Methods, 2007,4(8):613-614. |
| [28] | Buenrostro JD, Wu BJ, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-Wide. Curr Protoc Mol Biol, 2015, 109: 21.29.1-21.29.9. |
| [29] | Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ . Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods, 2013,10(12):1213-1218. |
| [30] | Schep AN, Buenrostro JD, Denny SK, Schwartz K, Sherlock G, Greenleaf WJ . Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res, 2015,25(11):1757-1770. |
| [31] | Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song LY, Vong S, Weaver M, Yan YQ, Zhang ZC, Zhang ZZ, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA . The accessible chromatin landscape of the human genome. Nature, 2012,489(7414):75-82. |
| [32] | Zhong JL, Luo KX, Winter PS, Crawford GE, Iversen ES, Hartemink AJ . Mapping nucleosome positions using DNase-seq. Genome Res, 2016,26(3):351-364. |
| [33] | Kelly TK, Liu YP, Lay FD, Liang GN, Berman BP, Jones PA . Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res, 2012,22(12):2497-2506. |
| [34] | Krebs AR, Imanci D, Hoerner L, Gaidatzis D, Burger L, Schübeler D. Genome-wide single-molecule footprinting reveals high RNA polymerase II turnover at paused promoters. Mol Cell, 2017,67(3): 411-422.e4. |
| [35] | Lai BB, Gao WW, Cui KR, Xie WL, Tang QS, Jin WF, Hu GQ, Ni B, Zhao KJ . Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature, 2018,562(7726):281-285. |
| [36] | Lion M, Tolstorukov MY, Oettinger MA . Low-Input MNase accessibility of chromatin (low-input MACC). Curr Protoc Mol Biol, 2019,127(1):e91. |
| [37] | Skene PJ, Henikoff JG, Henikoff S . Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc, 2018,13(5):1006-1019. |
| [38] | Hainer SJ, Bošković A, McCannell KN, Rando OJ, Fazzio TG. Profiling of pluripotency factors in single cells and early embryos. Cell, 2019, 177(5): 1319-1329.e11. |
| [39] | Gao WW, Lai BB, Ni B, Zhao KJ . Genome-wide profiling of nucleosome position and chromatin accessibility in single cells using scMNase-seq. Nat Protoc, 2020,15(1):68-85. |
| [40] | Baldi S . Nucleosome positioning and spacing: from genome-wide maps to single arrays. Essays Biochem, 2019,63(1):5-14. |
| [41] | Baldi S, Korber P, Becker PB . Beads on a string- nucleosome array arrangements and folding of the chromatin fiber. Nat Struct Mol Biol, 2020,27(2):109-118. |
| [42] | Mavrich TN, Jiang CZ, Ioshikhes IP, Li XY, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, Gilmour DS, Albert I, Pugh BF . Nucleosome organization in the Drosophila genome. Nature, 2008,453(7193):358-362. |
| [43] | Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C . A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet, 2007,39(10):1235-1244. |
| [44] | Schones DE, Cui KR, Cuddapah S, Roh TY, Barski A, Wang ZB, Wei G, Zhao KJ . Dynamic regulation of nucleosome positioning in the human genome. Cell, 2008,132(5):887-898. |
| [45] | Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res, 2008,18(7):1051-1063. |
| [46] | Lai WKM, Pugh BF . Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Biol, 2017,18(9):548-562. |
| [47] | Martin C, Zhang Y . Mechanisms of epigenetic inheritance. Curr Opin Cell Biol, 2007,19(3):266-272. |
| [48] | Ruthenburg AJ, Li HT, Patel DJ, Allis CD . Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol, 2007,8(12):983-994. |
| [49] | Ahmad K, Henikoff S . The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell, 2002,9(6):1191-1200. |
| [50] | Sarma K, Reinberg D . Histone variants meet their match. Nat Rev Mol Cell Biol, 2005,6(2):139-149. |
| [51] | Ramachandran S, Zentner GE, Henikoff S . Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res, 2015,25(3):381-390. |
| [52] | Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E,. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature, 2009,458(7236):362-366. |
| [53] | Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF . Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature, 2007,446(7135):572-576. |
| [54] | Cui F, Chen LL, LoVerso PR, Zhurkin VB. Prediction of nucleosome rotational positioning in yeast and human genomes based on sequence-dependent DNA anisotropy. BMC Bioinformatics, 2014,15(1):313. |
| [55] | de Dieuleveult M, Yen KY, Hmitou I, Depaux A, Boussouar F, Bou Dargham D, Jounier S, Humbertclaude H, Ribierre F, Baulard C, Farrell NP, Park B, Keime C, Carrière L, Berlivet S, Gut M, Gut L, Werner M, Deleuze JF, Olaso R, Aude JC, Chantalat S, Pugh BF, Gérard M . Genome-wide nucleosome specificity and function of chromatin remodellers in ES cells. Nature, 2016,530(7588):113-116. |
| [56] | Ho L, Crabtree GR . Chromatin remodelling during development. Nature, 2010,463(7280):474-484. |
| [57] | Dutta A, Gogol M, Kim JH, Smolle M, Venkatesh S, Gilmore J, Florens L, Washburn MP, Workman JL . Swi/Snf dynamics on stress-responsive genes is governed by competitive bromodomain interactions. Genes Dev, 2014,28(20):2314-2330. |
| [58] | Ribeiro-Silva C, Vermeulen W, Lans H . SWI/SNF: Complex complexes in genome stability and cancer. DNA Repair, 2019,77:87-95. |
| [59] | Aras S, Saladi SV, Basuroy T, Marathe HG, Lorès P, de la Serna IL,. BAF60A mediates interactions between the microphthalmia-associated transcription factor and the BRG1-containing SWI/SNF complex during melanocyte differentiation. J Cell Physiol, 2019,234(7):11780-11791. |
| [60] | Oppikofer M, Bai TY, Gan YT, Haley B, Liu P, Sandoval W, Ciferri C, Cochran AG . Expansion of the ISWI chromatin remodeler family with new active complexes. EMBO Rep, 2017,18(10):1697-1706. |
| [61] | Levendosky RF, Bowman GD . Asymmetry between the two acidic patches dictates the direction of nucleosome sliding by the ISWI chromatin remodeler. eLife, 2019,8:e45472. |
| [62] | Cairns BR . The logic of chromatin architecture and remodelling at promoters. Nature, 2009,461(7261):193-198. |
| [63] | Ocampo J, Chereji RV, Eriksson PR, Clark DJ . The ISW1 and CHD1 ATP-dependent chromatin remodelers compete to set nucleosome spacing in vivo. Nucleic Acids Res, 2016,44(10):4625-4635. |
| [64] | Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD . Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet, 2004,36(8):900-905. |
| [65] | Sekinger EA, Moqtaderi Z, Struhl K . Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell, 2005,18(6):735-748. |
| [66] | Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL . Global nucleosome occupancy in yeast. Genome Biol, 2004,5(9):R62. |
| [67] | Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ . Genome-scale identification of nucleosome positions in S. cerevisiae. Science, 2005,309(5734):626-630. |
| [68] | Rando OJ, Ahmad K . Rules and regulation in the primary structure of chromatin. Curr Opin Cell Biol, 2007,19(3):250-256. |
| [69] | Mavrich TN, Ioshikhes IP, Venters BJ, Jiang CZ, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF . A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res, 2008,18(7):1073-1083. |
| [70] | Kubik S, Bruzzone MJ, Challal D, Dreos R, Mattarocci S, Bucher P, Libri D, Shore D . Opposing chromatin remodelers control transcription initiation frequency and start site selection. Nat Struct Mol Biol, 2019,26(8):744-754. |
| [71] | Kubik S, O’Duibhir E, de Jonge WJ, Mattarocci S, Albert B, Falcone JL, Bruzzone MJ, Holstege FCP, Shore D. Sequence-directed action of RSC remodeler and general regulatory factors modulates +1 nucleosome position to facilitate transcription. Mol Cell, 2018, 71(1): 89-102.e5. |
| [72] | Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A . Determinants of nucleosome organization in primary human cells. Nature, 2011,474(7352):516-520. |
| [73] | Boeger H, Griesenbeck J, Strattan JS, Kornberg RD . Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell, 2004,14(5):667-673. |
| [74] | Ertel F, Dirac-Svejstrup AB, Hertel CB, Blaschke D, Svejstrup JQ, Korber P . In vitro reconstitution of PHO5 promoter chromatin remodeling points to a role for activator-nucleosome competition in vivo. Mol Cell Biol, 2010,30(16):4060-4076. |
| [75] | Shivaswamy S, Bhargava P . Positioned nucleosomes due to sequential remodeling of the yeast U6 small nuclear RNA chromatin are essential for its transcriptional activation. J Biol Chem, 2006,281(15):10461-10472. |
| [76] | Shivaswamy S, Bhinge A, Zhao YJ, Jones S, Hirst M, Iyer VR . Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol, 2008,6(3):e65. |
| [77] | Kulaeva OI, Hsieh FK, Chang HW, Luse DS, Studitsky VM . Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim Biophys Acta, 2013,1829(1):76-83. |
| [78] | Reja R, Vinayachandran V, Ghosh S, Pugh BF . Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev, 2015,29(18):1942-1954. |
| [79] | Lomvardas S, Thanos D . Modifying gene expression programs by altering core promoter chromatin architecture. Cell, 2002,110(2):261-271. |
| [80] | Ford E, Thanos D . The transcriptional code of human IFN-beta gene expression. Biochim Biophys Acta, 2010,1799(3-4):328-336. |
| [81] | Au-Yeung N, Horvath CM . Transcriptional and chromatin regulation in interferon and innate antiviral gene expression. Cytokine Growth Factor Rev, 2018,44:11-17. |
| [82] | Rothbart SB, Strahl BD . Interpreting the language of histone and DNA modifications. Biochim Biophys Acta, 2014,1839(8):627-643. |
| [83] | Felsenfeld G, Groudine M . Controlling the double helix. Nature, 2003,421(6921):448-453. |
| [84] | Qi HY, Zhang ZJ, Li YJ, Fang XD . Role of chromatin conformation in eukaryotic gene regulation. Hereditas (Beijing), 2011,33(12):1291-1299. |
| 亓合媛, 张昭军, 李雅娟, 方向东 . 染色质构象调控真核基因的表达. 遗传, 2011,33(12):1291-1299. | |
| [85] | Vogelauer M, Wu J, Suka N, Grunstein M . Global histone acetylation and deacetylation in yeast. Nature, 2000,408(6811):495-498. |
| [86] | Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL . Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci USA, 2002,99(13):8695-8700. |
| [87] | Klemm SL, Shipony Z, WJ. Chromatin accessibility and the regulatory epigenome. Nat Rev Genet, 2019,20(4):207-220. |
| [88] | Du YH, Liu ZP, Cao XK, Chen XL, Chen ZY, Zhang XB, Zhang XQ, Jiang CZ . Nucleosome eviction along with H3K9ac deposition enhances Sox2 binding during human neuroectodermal commitment. Cell Death Differ, 2017,24(6):1121-1131. |
| [89] | Daneshpajooh M, Bacos K, Bysani M, Bagge A, Ottosson Laakso E, Vikman P, Eliasson L, Mulder H, Ling C . HDAC7 is overexpressed in human diabetic islets and impairs insulin secretion in rat islets and clonal beta cells. Diabetologia, 2017,60(1):116-125. |
| [90] | Vallianatos CN, Raines B, Porter RS, Bonefas KM, Wu MC, Garay PM, Collette KM, Seo YA, Dou Y, Keegan CE, Tronson NC, Iwase S,. Mutually suppressive roles of KMT2A and KDM5C in behaviour, neuronal structure, and histone H3K4 methylation. Commun Biol, 2020,3(1):278. |
| [91] | Sasidharan Nair V, El Salhat H, Taha RZ, John A, Ali BR, Elkord E . DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin Epigenetics, 2018,10:78. |
| [92] | Barski A, Cuddapah S, Cui KR, Roh TY, Schones DE, Wang ZB, Wei G, Chepelev L, Zhao KJ . High-resolution profiling of histone methylations in the human genome. Cell, 2007,129(4):823-837. |
| [93] | Giaimo BD, Ferrante F, Herchenröther A, Hake SB, Borggrefe T . The histone variant H2A.Z in gene regulation. Epigenetics Chromatin, 2019,12(1):37. |
| [94] | Bagchi DN, Battenhouse AM, Park D, Iyer VR . The histone variant H2A.Z in yeast is almost exclusively incorporated into the +1 nucleosome in the direction of transcription. Nucleic Acids Res, 2020,48(1):157-170. |
| [95] | Ranjan A, Nguyen VQ, Liu S, Wisniewski J, Kim JM, Tang XN, Mizuguchi G, Elalaoui E, Nickels TJ, Jou V, English BP, Zheng QS, Luk E, Lavis LD, Lionnet T, Wu C,. Live-cell single particle imaging reveals the role of RNA polymerase II in histone H2A.Z eviction. eLife, 2020,9:e55667. |
| [96] | Murphy PJ, Wu SF, James CR, Wike CL, Cairns BR. Placeholder nucleosomes underlie Germline-to-Embryo DNA methylation reprogramming. Cell, 2018, 172(5): 993-1006.e13. |
| [97] | Sharma S, Kelly TK, Jones PA . Epigenetics in cancer. Carcinogenesis, 2010,31(1):27-36. |
| [98] | Wang HF, Fu C, Du J, Wang HS, He R, Yin XF, Li HX, Li X, Wang HX, Li K, Zheng L, Liu ZC, Qiu YR . Enhanced histone H3 acetylation of the PD-L1 promoter via the COP1/c-Jun/HDAC3 axis is required for PD-L1 expression in drug-resistant cancer cells. J Exp Clin Cancer Res, 2020,39(1):29. |
| [99] | Kumar A, Kumari N, Sharma U, Ram S, Singh SK, Kakkar N, Kaushal K, Prasad R . Reduction in H3K4me patterns due to aberrant expression of methyltransferases and demethylases in renal cell carcinoma: prognostic and therapeutic implications. Sci Rep, 2019,9(1):8189. |
| [100] | Webber LP, Wagner VP, Curra M, Vargas PA, Meurer L, Carrard VC, Squarize CH, Castilho RM, Martins MD . Hypoacetylation of acetyl-histone H3 (H3K9ac) as marker of poor prognosis in oral cancer. Histopathology, 2017,71(2):278-286. |
| [101] | Torres CM, Biran A, Burney MJ, Patel H, Henser- Brownhill T, Cohen AHS, Li YL, Ben-Hamo R, Nye E, Spencer-Dene B, Chakravarty P, Efroni S, Matthews N, Misteli T, Meshorer E, Scaffidi P. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science, 2016, 353(6307): aaf1644. |
| [102] | Huang QT, Li Q, Zhang YB . Linking chromatin conformation to gene function. Hereditas(Beijing), 2020,42(1):1-17. |
| 黄其通, 李清, 张玉波 . 染色质构象与基因功能. 遗传, 2020,42(1):1-17. | |
| [103] | Denker A, de Laat W . The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev, 2016,30(12):1357-1382. |
| [1] | Shunze Wang, Feng Jiang, Dongli Zhu, Tie-Lin Yang, Yan Guo. Application of Hi-C technology in three-dimensional genomics research and disease pathogenesis analysis [J]. Hereditas(Beijing), 2023, 45(4): 279-294. |
| [2] | Haoliang Cui, Peihua Shi, Jinchun Gao, Xinbo Zhang, Shunran Zhao, Chenyu Tao. Progress on the study of nucleosome reorganization during cellular reprogramming [J]. Hereditas(Beijing), 2022, 44(3): 208-215. |
| [3] | Jingwen Zhang,Qian Xu,Guoliang Li. Epigenetics in the genesis and development of cancers [J]. Hereditas(Beijing), 2019, 41(7): 567-581. |
| [4] | Hui Zhao, Yongchao Zhang, Yongqing Zhang. Recent progresses in molecular genetics of autism spectrum disorders [J]. HEREDITAS(Beijing), 2015, 37(9): 845-854. |
| [5] | Jinxuan Zhao, Fang Wang, Zhengrong Xu, Yimei Fan. The epigenetic effect on pre-mRNA alternative splicing [J]. HEREDITAS, 2014, 36(3): 248-255. |
| [6] | Jiapeng Zhou, Zhiyong Pei, Yubao Chen, Runsheng Chen. Strategies of genome-wide association study based on high-throughput sequencing [J]. HEREDITAS(Beijing), 2014, 36(11): 1099-1111. |
| [7] | WANG Ting-Zhang, SHAN Gao, XU Jian-Hong, XUE Qing-Zhong. Genome-scale sequence data processing and epigenetic analysis of DNA methylation [J]. HEREDITAS, 2013, 35(6): 685-684. |
| [8] | QI Yun-Xia, LIU Yong-Bin, RONG Wei-Heng. RNA-Seq and its applications: a new technology for transcriptomics [J]. HEREDITAS, 2011, 33(11): 1191-1202. |
| [9] | WANG Juan, CENG Xian-Lu. ATP-dependent chromatin remodeling complex and its function in regulating chromatin structure [J]. HEREDITAS, 2010, 32(4): 301-306. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||