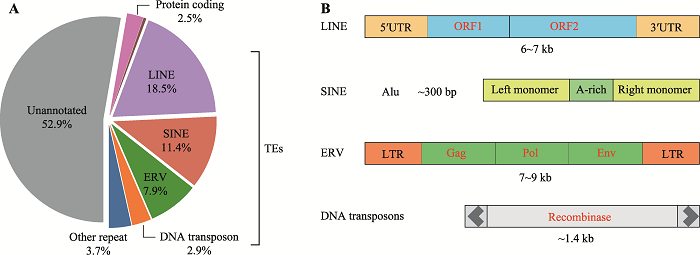
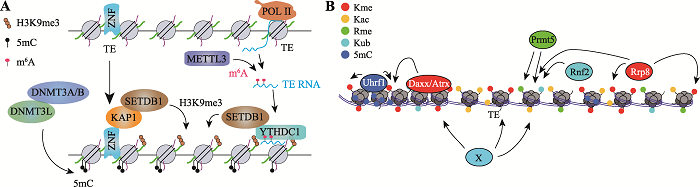
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (9): 822-834.doi: 10.16288/j.yczz.21-113
• Special Section: Excellent Doctoral Thesis • Previous Articles Next Articles
Epigenetic control of transposable elements and cell fate decision
Jiangping He1,2, Jiekai Chen1,2( )
)
- 1. Center for Cell Lineage and Atlas (CCLA), Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510530, China
2. Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China;
-
Received:2021-03-25Revised:2021-07-23Online:2021-09-20Published:2021-08-19 -
Contact:Chen Jiekai E-mail:chen_jiekai@gibh.ac.cn -
Supported by:Supported by the National Key R&D Program of China No(2019YFA0110200);the Frontier Science Research Program of the CAS No(ZDBS-LY-SM007)
Cite this article
Jiangping He, Jiekai Chen. Epigenetic control of transposable elements and cell fate decision[J]. Hereditas(Beijing), 2021, 43(9): 822-834.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
McCLINTOCK B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol, 1951, 16:13-47.
pmid: 14942727 |
| [2] | Cui XK, Cao XF. Overview of the function of transposable elements in higher plants. Prog Biochem Biophys, 2015, 42(11):1033-1046. |
| 崔勰奎, 曹晓风. 高等植物转座元件功能研究进展. 生物化学与生物物理进展, 2015, 42(11):1033-1046. | |
| [3] | Hutchins AP, Pei DQ. Transposable elements at the center of the crossroads between embryogenesis, embryonic stem cells, reprogramming, and long non-coding RNAs. Sci Bull (Beijing), 2015, 60(20):1722-1733. |
| [4] |
Pace JK, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res, 2007, 17(4):422-432.
doi: 10.1101/gr.5826307 |
| [5] |
Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet, 2011, 12:187-215.
doi: 10.1146/annurev-genom-082509-141802 |
| [6] |
Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet, 2009, 10(10):691-703.
doi: 10.1038/nrg2640 |
| [7] |
Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature, 2014, 516(7530):242-245.
doi: 10.1038/nature13760 |
| [8] | Wang JL, Wang J, Tian CY. Evolution of KRAB- containing zinc finger proteins and their roles in species evolution. Hereditas(Beijing), 2016, 38(11):971-978. |
| 王进龙, 王建, 田春艳. KRAB型锌指蛋白的进化及在物种演化中的功能. 遗传, 2016, 38(11):971-978. | |
| [9] |
Imbeault M, Helleboid PY, Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature, 2017, 543(7646):550-554.
doi: 10.1038/nature21683 |
| [10] |
Silva JC, Shabalina SA, Harris DG, Spouge JL, Kondrashovi AS. Conserved fragments of transposable elements in intergenic regions: evidence for widespread recruitment of MIR- and L2-derived sequences within the mouse and human genomes. Genet Res, 2003, 82(1):1-18.
doi: 10.1017/S0016672303006268 |
| [11] |
Friedli M, Trono D. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol, 2015, 31:429-451.
doi: 10.1146/annurev-cellbio-100814-125514 pmid: 26393776 |
| [12] |
Xie XH, Kamal M, Lander ES. A family of conserved noncoding elements derived from an ancient transposable element. Proc Natl Acad Sci USA, 2006, 103(31):11659-11664.
doi: 10.1073/pnas.0604768103 |
| [13] |
Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, Jurka J, Kamal M, Mauceli E, Searle SMJ, Sharpe T, Baker ML, Batzer MA, Benos PV, Belov K, Clamp M, Cook A, Cuff J, Das R, Davidow L, Deakin JE, Fazzari MJ, Glass JL, Grabherr M, Greally JM, Gu WJ, Hore TA, Huttley GA, Kleber M, Jirtle RL, Koina E, Lee JT, Mahony S, Marra MA, Miller RD, Nicholls RD, Oda M, Papenfuss AT, Parra ZE, Pollock DD, Ray DA, Schein JE, Speed TP, Thompson K, VandeBerg JL, Wade CM, Walker JA, Waters PD, Webber C, Weidman JR, Xie XH, Zody MC, Broad Institute Genome Sequencing Platform; Broad Institute Whole Genome Assembly Team; Graves JAM, Ponting CP, Breen M, Samollow PB, Lander ES, Lindblad-Toh K. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature, 2007, 447(7141):167-177.
doi: 10.1038/nature05805 |
| [14] | Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet, 2017, 18(2):71-86. |
| [15] |
Groh S, Schotta G. Silencing of endogenous retroviruses by heterochromatin. Cell Mol Life Sci, 2017, 74(11):2055-2065.
doi: 10.1007/s00018-017-2454-8 |
| [16] |
Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, Lorincz MC. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell, 2011, 8(6):676-687.
doi: 10.1016/j.stem.2011.04.004 |
| [17] |
Rowe HM, Trono D. Dynamic control of endogenous retroviruses during development. Virology, 2011, 411(2):273-287.
doi: 10.1016/j.virol.2010.12.007 |
| [18] |
Tan XY, Xu XB, Elkenani M, Smorag L, Zechner U, Nolte J, Engel W, Pantakani DVK. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res, 2013, 11(3):1045-1059.
doi: 10.1016/j.scr.2013.07.006 |
| [19] |
Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, Spitz F, Constam DB, Trono D. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature, 2010, 463(7278):237-240.
doi: 10.1038/nature08674 |
| [20] |
Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature, 2009, 458(7242):1201-1204.
doi: 10.1038/nature07844 |
| [21] |
Schoorlemmer J, Pérez-Palacios R, Climent M, Guallar D, Muniesa P. Regulation of mouse retroelement MuERV-L/ MERVL expression by REX1 and epigenetic control of stem cell potency. Front Oncol, 2014, 4:14.
doi: 10.3389/fonc.2014.00014 pmid: 24567914 |
| [22] |
Yang BX, Farran CAE, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YFS, Goh GYL, Neo SP, Li YH, Lorincz MC, Tergaonkar V, Lim TM, Chen LY, Gunaratne J, Collins JJ, Goff SP, Daley GQ, Li H, Bard FA, Loh YH. Systematic identification of factors for provirus silencing in embryonic stem cells. Cell, 2015, 163(1):230-245.
doi: 10.1016/j.cell.2015.08.037 pmid: 26365490 |
| [23] |
Bulut-Karslioglu A, De La Rosa-Velázquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galán C, Winter GE, Engist B, Gerle B, O'Sullivan RJ, Martens JHA, Walter J, Manke T, Lachner M, Jenuwein T. Suv39h- dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell, 2014, 55(2):277-290.
doi: 10.1016/j.molcel.2014.05.029 pmid: 24981170 |
| [24] |
He QY, Kim H, Huang R, Lu WS, Tang MF, Shi FT, Yang D, Zhang XY, Huang JJ, Liu D, Zhou SY. The Daxx/Atrx complex protects tandem repetitive elements during DNA hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell, 2015, 17(3):273-286.
doi: 10.1016/j.stem.2015.07.022 |
| [25] |
Xu WQ, Li JH, He CX, Wen J, Ma HH, Rong BW, Diao JB, Wang LY, Wang JH, Wu FZ, Tan L, Shi YG, Shi Y, Shen HJ. METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature, 2021, 591(7849):317-321.
doi: 10.1038/s41586-021-03210-1 |
| [26] |
Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, Dossin F, Fouassier C, Lameiras S, Bourc'his D. m6 A RNA methylation regulates the fate of endogenous retroviruses. Nature, 2021, 591(7849):312-316.
doi: 10.1038/s41586-020-03135-1 |
| [27] |
Liu J, Dou XY, Chen CY, Chen C, Liu C, Xu MM, Zhao SQ, Shen B, Gao YW, Han DL, He C. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science, 2020, 367(6477):580-586.
doi: 10.1126/science.aay6018 |
| [28] |
Elsässer SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature, 2015, 522(7555):240-244.
doi: 10.1038/nature14345 |
| [29] |
Kim S, Günesdogan U, Zylicz JJ, Hackett JA, Cougot D, Bao S, Lee C, Dietmann S, Allen GE, Sengupta R, Surani MA. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol Cell, 2014, 56(4):564-579.
doi: 10.1016/j.molcel.2014.10.003 |
| [30] |
Esnault C, Heidmann O, Delebecque F, Dewannieux M, Ribet D, Hance AJ, Heidmann T, Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature, 2005, 433(7024):430-433.
doi: 10.1038/nature03238 |
| [31] |
Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. eLife, 2014, 3:e02008.
doi: 10.7554/eLife.02008 |
| [32] |
Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res, 2006, 34(5):1522-1531.
pmid: 16537839 |
| [33] |
Isbel L, Srivastava R, Oey H, Spurling A, Daxinger L, Puthalakath H, Whitelaw E. Trim33 binds and silences a class of young endogenous retroviruses in the mouse testis; a novel component of the arms race between retrotransposons and the host genome. PLoS Genet, 2015, 11(12):e1005693.
doi: 10.1371/journal.pgen.1005693 |
| [34] |
Ancelin K, Syx L, Borensztein M, Ranisavljevic N, Vassilev I, Briseño-Roa L, Liu T, Metzger E, Servant N, Barillot E, Chen CJ, Schüle R, Heard E. Maternal LSD1/KDM1A is an essential regulator of chromatin and transcription landscapes during zygotic genome activation. eLife, 2016, 5:e08851.
doi: 10.7554/eLife.08851 |
| [35] |
Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol, 2015, 22(9):662-671.
doi: 10.1038/nsmb.3066 pmid: 26237512 |
| [36] |
Li PS, Wang L, Bennett BD, Wang JJ, Li JL, Qin YF, Takaku M, Wade PA, Wong JM, Hu G. Rif1 promotes a repressive chromatin state to safeguard against endogenous retrovirus activation. Nucleic Acids Res, 2017, 45(22):12723-12738.
doi: 10.1093/nar/gkx884 |
| [37] |
Guallar D, Bi XJ, Pardavila JA, Huang X, Saenz C, Shi XL, Zhou HW, Faiola F, Ding JJ, Haruehanroengra P, Yang F, Li D, Sanchez-Priego C, Saunders A, Pan F, Valdes VJ, Kelley K, Blanco MG, Chen LY, Wang HY, Sheng J, Xu MJ, Fidalgo M, Shen XH, Wang JL. RNA-dependent chromatin targeting of TET2 for endogenous retrovirus control in pluripotent stem cells. Nat Genet, 2018, 50(3):443-451.
doi: 10.1038/s41588-018-0060-9 pmid: 29483655 |
| [38] |
He JP, Fu XL, Zhang M, He FF, Li WJ, Abdul MM, Zhou JG, Sun L, Chang C, Li YH, Liu H, Wu KX, Babarinde IA, Zhuang Q, Loh YH, Chen JK, Esteban MA, Hutchins AP. Transposable elements are regulated by context-specific patterns of chromatin marks in mouse embryonic stem cells. Nat Commun, 2019, 10(1):34.
doi: 10.1038/s41467-018-08006-y |
| [39] |
Chuong EB, Rumi MAK, Soares MJ, Baker JC. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet, 2013, 45(3):325-329.
doi: 10.1038/ng.2553 |
| [40] |
Xie MC, Hong CB, Zhang B, Lowdon RF, Xing XY, Li DF, Zhou X, Lee HJ, Maire CL, Ligon KL, Gascard P, Sigaroudinia M, Tlsty TD, Kadlecek T, Weiss A, O'Geen H, Farnham PJ, Madden PAF, Mungall AJ, Tam A, Kamoh B, Cho S, Moore R, Hirst M, Marra MA, Costello JF, Wang T. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet, 2013, 45(7):836-841.
doi: 10.1038/ng.2649 |
| [41] |
Sundaram V, Choudhary MNK, Pehrsson E, Xing XY, Fiore C, Pandey M, Maricque B, Udawatta M, Ngo D, Chen YJ, Paguntalan A, Ray T, Hughes A, Cohen BA, Wang T. Functional cis-regulatory modules encoded by mouse-specific endogenous retrovirus. Nat Commun, 2017, 8:14550.
doi: 10.1038/ncomms14550 pmid: 28348391 |
| [42] |
Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvák Z, Levin HL, Macfarlan TS, Mager DL, Feschotte C. Ten things you should know about transposable elements. Genome Biol, 2018, 19(1):199.
doi: 10.1186/s13059-018-1577-z |
| [43] |
Wei LY, Cao XF. The effect of transposable elements on phenotypic variation: insights from plants to humans. Sci China Life Sci, 2016, 59(1):24-37.
doi: 10.1007/s11427-015-4993-2 |
| [44] |
Jordan IK, Rogozin IB, Glazko GV, Koonin EV. Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet, 2003, 19(2):68-72.
pmid: 12547512 |
| [45] |
Jjingo D, Huda A, Gundapuneni M, Mariño-Ramírez L, Jordan IK. Effect of the transposable element environment of human genes on gene length and expression. Genome Biol Evol, 2011, 3:259-271.
doi: 10.1093/gbe/evr015 pmid: 21362639 |
| [46] |
Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science, 2016, 351(6277):1083-1087.
doi: 10.1126/science.aad5497 |
| [47] |
Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu XY, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet, 2010, 42(7):631-634.
doi: 10.1038/ng.600 pmid: 20526341 |
| [48] |
Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science, 2016, 351(6274): aac7247.
doi: 10.1126/science.aac7247 |
| [49] |
Chen LL, DeCerbo JN, Carmichael GG. Alu element- mediated gene silencing. EMBO J, 2008, 27(12):1694-1705.
doi: 10.1038/emboj.2008.94 |
| [50] |
Feschotte C. Transposable elements and the evolution of regulatory networks. Nat Rev Genet, 2008, 9(5):397-405.
doi: 10.1038/nrg2337 pmid: 18368054 |
| [51] |
Ito J, Sugimoto R, Nakaoka H, Yamada S, Kimura T, Hayano T, Inoue I. Systematic identification and characterization of regulatory elements derived from human endogenous retroviruses. PLoS Genet, 2017, 13(7):e1006883.
doi: 10.1371/journal.pgen.1006883 |
| [52] |
Lu JY, Chang L, Li T, Wang T, Yin YF, Zhan G, Han X, Zhang K, Tao YB, Percharde M, Wang L, Peng Q, Yan PX, Zhang H, Bi XJ, Shao W, Hong YT, Wu ZY, Ma RZ, Wang PZ, Li WZ, Zhang J, Chang Z, Hou YP, Zhu B, Ramalho-Santos M, Li PL, Xie W, Na J, Sun YJ, Shen XH. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res, 2021, 31(6):613-630.
doi: 10.1038/s41422-020-00466-6 |
| [53] |
Kruse K, Díaz N, Enriquez-Gasca R, Gaume X, Torres- Padilla ME, Vaquerizas JM. Transposable elements drive reorganisation of 3D chromatin during early embryogenesis. bioRxiv, 2019, doi: https://doi.org/10.1101/523712.
doi: https://doi.org/10.1101/523712 |
| [54] |
Zhang YX, Li T, Preissl S, Amaral ML, Grinstein JD, Farah EN, Destici E, Qiu YJ, Hu R, Lee AY, Chee S, Ma KY, Ye Z, Zhu Q, Huang H, Fang RX, Yu LQ, Belmonte JCI, Wu J, Evans SM, Chi NC, Ren B. Transcriptionally active HERV-H retrotransposons demarcate topologically associating domains in human pluripotent stem cells. Nat Genet, 2019, 51(9):1380-1388.
doi: 10.1038/s41588-019-0479-7 |
| [55] |
Diehl AG, Ouyang NX, Boyle AP. Transposable elements contribute to cell and species-specific chromatin looping and gene regulation in mammalian genomes. Nat Commun, 2020, 11(1):1796.
doi: 10.1038/s41467-020-15520-5 pmid: 32286261 |
| [56] |
Cao YQ, Chen GY, Wu G, Zhang XL, McDermott J, Chen XW, Xu C, Jiang QL, Chen ZX, Zeng YY, Ai DS, Huang Y, Han JDJ. Widespread roles of enhancer-like transposable elements in cell identity and long-range genomic interactions. Genome Res, 2019, 29(1):40-52.
doi: 10.1101/gr.235747.118 |
| [57] |
Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell, 2012, 148(1-2):335-348.
doi: 10.1016/j.cell.2011.11.058 |
| [58] |
Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol, 2004, 24(6):2478-2486.
doi: 10.1128/MCB.24.6.2478-2486.2004 |
| [59] |
Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 1999, 99(3):247-257.
pmid: 10555141 |
| [60] |
Andricovich J, Kai Y, Peng WQ, Foudi A, Tzatsos A. Histone demethylase KDM2B regulates lineage commitment in normal and malignant hematopoiesis. J Clin Invest, 2016, 126(3):905-920.
doi: 10.1172/JCI84014 pmid: 26808549 |
| [61] |
Kawakami E, Tokunaga A, Ozawa M, Sakamoto R, Yoshida N. The histone demethylase Fbxl11/Kdm2a plays an essential role in embryonic development by repressing cell-cycle regulators. Mech Dev, 2015, 135:31-42.
doi: 10.1016/j.mod.2014.10.001 |
| [62] |
Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun WT, Chang H, Xu GL, Gaudet F, Li E, Chen TP. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet, 2009, 41(1):125-129.
doi: 10.1038/ng.268 |
| [63] |
Chen JK, Liu H, Liu J, Qi J, Wei B, Yang JQ, Liang HQ, Chen Y, Chen J, Wu YR, Guo L, Zhu JY, Zhao XJ, Peng TR, Zhang YX, Chen S, Li XJ, Li DW, Wang T, Pei DQ. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet, 2013, 45(1):34-42.
doi: 10.1038/ng.2491 |
| [64] |
Chen JK, Guo L, Zhang L, Wu HY, Yang JQ, Liu H, Wang XS, Hu X, Gu TP, Zhou ZW, Liu J, Liu JD, Wu HL, Mao SQ, Mo KL, Li YY, Lai KY, Qi J, Yao HJ, Pan GJ, Xu GL, Pei DQ. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet, 2013, 45(12):1504-1509.
doi: 10.1038/ng.2807 |
| [65] |
Sardina JL, Collombet S, Tian TV, Gómez A, Di Stefano B, Berenguer C, Brumbaugh J, Stadhouders R, Segura- Morales C, Gut M, Gut IG, Heath S, Aranda S, Di Croce L, Hochedlinger K, Thieffry D, Graf T. Transcription factors drive Tet2-mediated enhancer demethylation to reprogram cell fate. Cell Stem Cell, 2018, 23(5): 727-741.e9.
doi: S1934-5909(18)30401-6 pmid: 30220521 |
| [66] |
Cheloufi S, Elling U, Hopfgartner B, Jung YL, Murn J, Ninova M, Hubmann M, Badeaux AI, Ang CE, Tenen D, Wesche DJ, Abazova N, Hogue M, Tasdemir N, Brumbaugh J, Rathert P, Jude J, Ferrari F, Blanco A, Fellner M, Wenzel D, Zinner M, Vidal SE, Bell O, Stadtfeld M, Chang HY, Almouzni G, Lowe SW, Rinn J, Wernig M, Aravin A, Shi Y, Park PJ, Penninger JM, Zuber J, Hochedlinger K. The histone chaperone CAF-1 safeguards somatic cell identity. Nature, 2015, 528(7581):218-224.
doi: 10.1038/nature15749 |
| [67] |
Zhuang Q, Li WJ, Benda C, Huang ZJ, Ahmed T, Liu P, Guo XP, Ibañez DP, Luo ZW, Zhang M, Abdul MM, Yang ZZ, Yang JY, Huang YH, Zhang H, Huang DH, Zhou JG, Zhong XF, Zhu XH, Fu XL, Fan WX, Liu YL, Xu Y, Ward C, Khan MJ, Kanwal S, Mirza B, Tortorella MD, Tse HF, Chen JY, Qin BM, Bao XC, Gao SR, Hutchins AP, Esteban MA. NCoR/SMRT co-repressors cooperate with c-MYC to create an epigenetic barrier to somatic cell reprogramming. Nat Cell Biol, 2018, 20(4):400-412.
doi: 10.1038/s41556-018-0047-x pmid: 29531310 |
| [68] |
Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, Lander ES, Armstrong SA, Daley GQ. Chromatin-modifying enzymes as modulators of reprogramming. Nature, 2012, 483(7391):598-602.
doi: 10.1038/nature10953 |
| [69] |
Li HH, Lai P, Jia JP, Song YW, Xia Q, Huang KM, He N, Ping WF, Chen JY, Yang ZZ, Li J, Yao MZ, Dong XT, Zhao JC, Hou CH, Esteban MA, Gao SR, Pei DQ, Hutchins AP, Yao HJ. RNA helicase DDX5 inhibits reprogramming to pluripotency by miRNA-based repression of RYBP and its PRC1-dependent and -independent functions. Cell Stem Cell, 2017, 20(4): 462-477.e6.
doi: 10.1016/j.stem.2016.12.002 |
| [70] |
Zhou ZW, Yang XJ, He JP, Liu J, Wu F, Yu SY, Liu YT, Lin RX, Liu H, Cui YB, Zhou CH, Wang XS, Wu J, Cao ST, Guo L, Lin LH, Wang T, Peng XZ, Qiang BQ, Hutchins AP, Pei DQ, Chen JK. Kdm2b regulates somatic reprogramming through variant PRC1 complex-dependent function. Cell Rep, 2017, 21(8):2160-2170.
doi: 10.1016/j.celrep.2017.10.091 |
| [71] |
Wang T, Chen KS, Zeng XM, Yang JG, Wu Y, Shi X, Qin BM, Zeng LW, Esteban MA, Pan GJ, Pei DQ. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell, 2011, 9(6):575-587.
doi: 10.1016/j.stem.2011.10.005 pmid: 22100412 |
| [72] | Smith ZD, Sindhu C, Meissner A. Molecular features of cellular reprogramming and development. Nat Rev Mol Cell Biol, 2016, 17(3):139-154. |
| [73] |
Hörmanseder E, Simeone A, Allen GE, Bradshaw CR, Figlmüller M, Gurdon J, Jullien J. H3K4 methylation- dependent memory of somatic cell identity inhibits reprogramming and development of nuclear transfer embryos. Cell Stem Cell, 2017, 21(1): 135-143.e6.
doi: S1934-5909(17)30074-7 pmid: 28366589 |
| [74] | Deniz Ö, Frost JM, Branco MR. Regulation of transposable elements by DNA modifications. Nat Rev Genet, 2019, 20(7):417-431. |
| [75] |
Percharde M, Lin CJ, Yin YF, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen XH, Ramalho-Santos M. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell, 2018, 174(2): 391-405.e19.
doi: S0092-8674(18)30655-X pmid: 29937225 |
| [76] |
Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature, 2012, 487(7405):57-63.
doi: 10.1038/nature11244 |
| [77] |
Jachowicz JW. Bing XY, Pontabry J, Bošković A, Rando OJ, Torres-Padilla ME. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet, 2017, 49(10):1502-1510.
doi: 10.1038/ng.3945 pmid: 28846101 |
| [78] |
Muotri AR, Chu VT, Marchetto MCN, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature, 2005, 435(7044):903-910.
doi: 10.1038/nature03663 |
| [79] |
Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu YL, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature, 2009, 460(7259):1127-1131.
doi: 10.1038/nature08248 |
| [80] |
Friedli M, Turelli P, Kapopoulou A, Rauwel B, Castro- Díaz N, Rowe HM, Ecco G, Unzu C, Planet E, Lombardo A, Mangeat B, Wildhaber BE, Naldini L, Trono D. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res, 2014, 24(8):1251-1259.
doi: 10.1101/gr.172809.114 pmid: 24879558 |
| [81] |
Theunissen TW, Friedli M, He YP, Planet E, O'Neil RC, Markoulaki S, Pontis J, Wang HY, Iouranova A, Imbeault M, Duc J, Cohen MA, Wert KJ, Castanon R, Zhang ZZ, Huang YM, Nery JR, Drotar J, Lungjangwa T, Trono D, Ecker JR, Jaenisch R. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell, 2016, 19(4):502-515.
doi: S1934-5909(16)30161-8 pmid: 27424783 |
| [82] |
Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, Pera RAR, Wysocka J. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature, 2015, 522(7555):221-225.
doi: 10.1038/nature14308 |
| [83] |
Wang JC, Xie GC, Singh M, Ghanbarian AT, Raskó T, Szvetnik A, Cai HQ, Besser D, Prigione A, Fuchs NV, Schumann GG, Chen W, Lorincz MC, Ivics Z, Hurst LD, Izsvák Z. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature, 2014, 516(7531):405-409.
doi: 10.1038/nature13804 |
| [84] |
Ohnuki M, Tanabe K, Sutou K, Teramoto I, Sawamura Y, Narita M, Nakamura M, Tokunaga Y, Nakamura M, Watanabe A, Yamanaka S, Takahashi K. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc Natl Acad Sci USA, 2014, 111(34):12426-12431.
doi: 10.1073/pnas.1413299111 |
| [85] |
Adoue V, Binet B, Malbec A, Fourquet J, Romagnoli P, van Meerwijk JPM, Amigorena S, Joffre OP. The histone methyltransferase SETDB1 controls T helper cell lineage integrity by repressing endogenous retroviruses. Immunity, 2019, 50(3): 629-644.e8.
doi: 10.1016/j.immuni.2019.01.003 |
| [86] |
Goerner-Potvin P, Bourque G. Computational tools to unmask transposable elements. Nat Rev Genet, 2018, 19(11):688-704.
doi: 10.1038/s41576-018-0050-x pmid: 30232369 |
| [87] |
Treangen TJ, Salzberg SL. Repetitive DNA and next- generation sequencing: computational challenges and solutions. Nat Rev Genet, 2011, 13(1):36-46.
doi: 10.1038/nrg3117 pmid: 22124482 |
| [88] |
Nellåker C, Keane TM, Yalcin B, Wong K, Agam A, Belgard TG, Flint J, Adams DJ, Frankel WN, Ponting CP. The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol, 2012, 13(6):R45.
doi: 10.1186/gb-2012-13-6-r45 |
| [89] | He JP, Babarinde IA, Sun L, Xu SY, Chen RH, Wei YJ, Li YH, Ma G, Zhuang Q, Hutchins AP, Chen JK. Unveiling transposable element expression heterogeneity in cell fate regulation at the single-cell level. bioRxiv, 2020, https://doi.org/10.1101/2020.07.23.218800. |
| [90] |
Shao WQ, Wang T. Transcript assembly improves expression quantification of transposable elements in single-cell RNA-seq data. Genome Res, 2021, 31(1):88-100.
doi: 10.1101/gr.265173.120 |
| [91] |
Niu D, Wei HJ, Lin L, George H, Wang T, Lee IH, Zhao HY, Wang Y, Kan YN, Shrock E, Lesha E, Wang G, Luo YL, Qing YB, Jiao DL, Zhao H, Zhou XY, Wang SQ, Wei H, Güell M, Church GM, Yang LH. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science, 2017, 357(6357):1303-1307.
doi: 10.1126/science.aan4187 |
| [92] |
Fuentes DR, Swigut T, Wysocka J. Systematic perturbation of retroviral LTRs reveals widespread long-range effects on human gene regulation. eLife, 2018, 7:e35989.
doi: 10.7554/eLife.35989 |
| [1] | Yan Zhao, Chenxin Wang, Tianming Yang, Chunshuang Li, Lihong Zhang, Dongni Du, Ruoxi Wang, Jing Wang, Min Wei, Xueqing Ba. Linking oxidative DNA lesion 8-OxoG to tumor development and progression [J]. Hereditas(Beijing), 2022, 44(6): 466-477. |
| [2] | Xiangrong Cheng,Xinglin Hu,Qi Jiang,Xingwei Huang,Nan Wang,Lei Lei. The epigenetic regulation of ribosomal DNA and tumorigenesis [J]. Hereditas(Beijing), 2019, 41(3): 185-192. |
| [3] | Jian Wang, Kaixiang Zhang, Guozhen Lu, Xianghui Zhao. Research progress on 5hmC and TET dioxygenases in neurodevelopment and neurological diseases [J]. Hereditas(Beijing), 2017, 39(12): 1138-1149. |
| [4] | Tingting Cui, Tianyu Xing, Yankan Chu, Hui Li, Ning Wang. Genetic and epigenetic regulation of PPARγ during adipogenesis [J]. Hereditas(Beijing), 2017, 39(11): 1066-1077. |
| [5] | Lili Yu,Wanru Dong,Minghui Chen,Xiangyang Kong. Progress in the molecular genetic mechanism of gonadoblastoma [J]. HEREDITAS(Beijing), 2015, 37(11): 1105-1115. |
| [6] | XU Hong-En, ZHANG Hua-Hao, HAN Min-Jin, SHEN Yi-Hong, HUANG Xian-Zhi, XIANG Zhong-Huai, ZHANG Ze. Computational approaches for identification and classification of transposable elements in eukaryotic genomes [J]. HEREDITAS, 2012, 34(8): 1009-1019. |
| [7] | GE Shao-Qin, LI Jian-Zhong, ZHANG Xiao-Jing. Methylation and acetylation of histones during spermatogenesis [J]. HEREDITAS, 2011, 33(9): 939-946. |
| [8] | FENG Bi-Wei, CHEN Jian-Jiang, LEI Bing-Kun, BO Xian, LV Gong. Research progress on genomic integrity regulated by epigenetics us-ing yeast as a model [J]. HEREDITAS, 2010, 32(8): 799-807. |
| [9] | CHENG Wei, MA Duan. Research progress in modification of histone lysine methylation and congenital heart defect [J]. HEREDITAS, 2010, 32(7): 650-655. |
| [10] | CHEN Jian-Jun, WANG Ying. Recent progress in plant genome size evolution [J]. HEREDITAS, 2009, 31(5): 464-464―470. |
| [11] | ZHUANG Jun, LIU Zhi-Xin. The Evolution of Plant Disease Resistance Gene [J]. HEREDITAS, 2004, 26(6): 962-968. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||