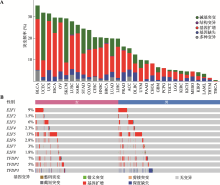
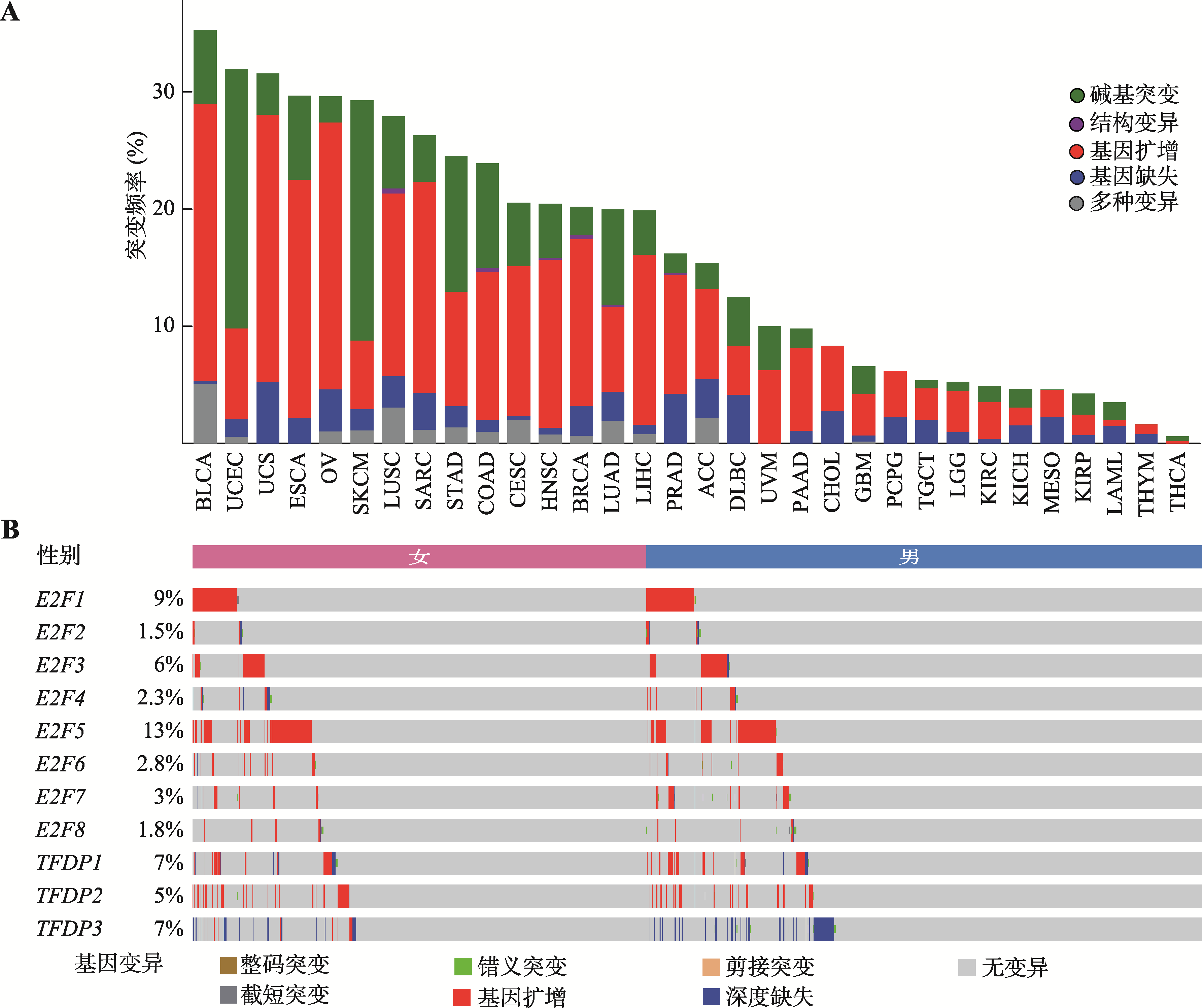
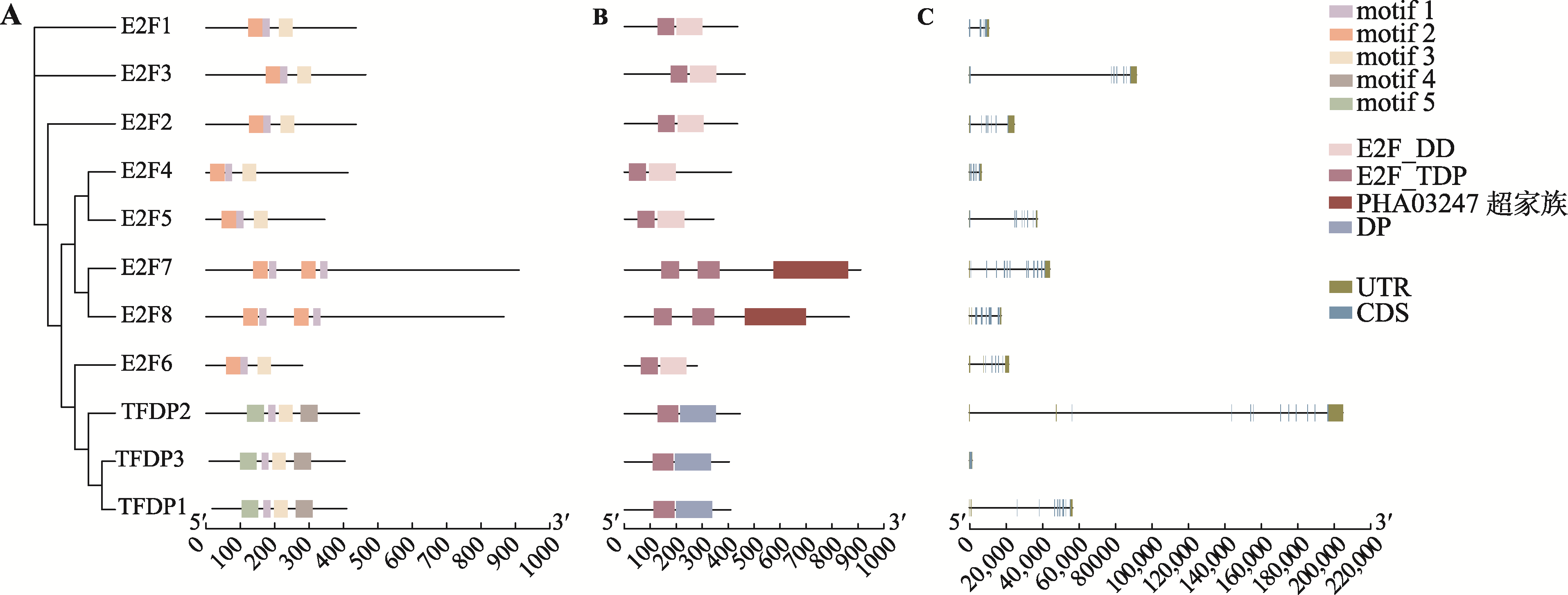
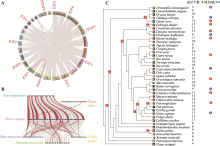
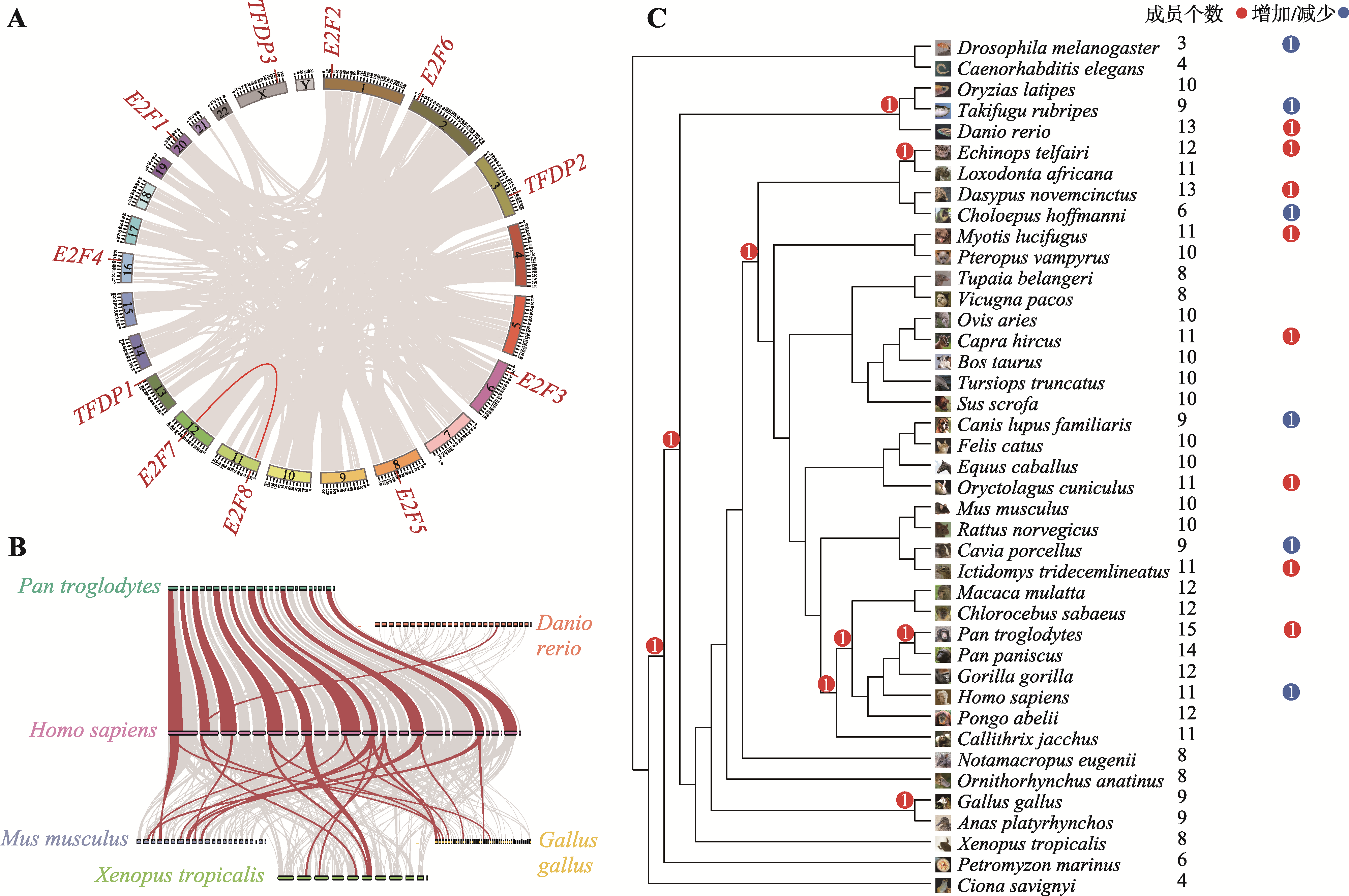
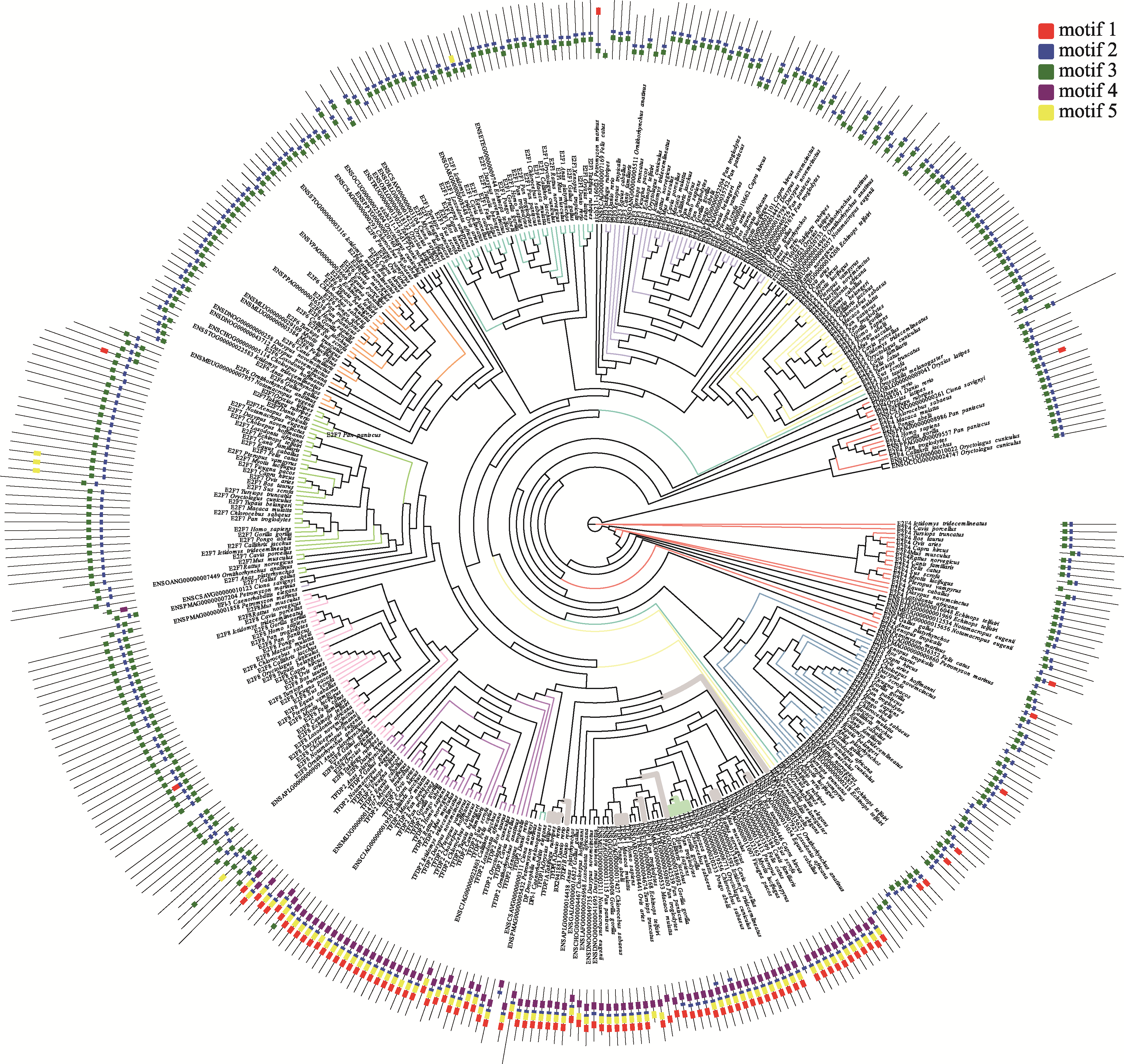
Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (7): 580-592.doi: 10.16288/j.yczz.23-029
• Research Article • Previous Articles Next Articles
E2F family play important roles in tumorigenesis
Feifei Li1( ), Yun Wang1, Jihai Gu1,2, Yuming Zhang1,2, Fengsong Liu1,2(
), Yun Wang1, Jihai Gu1,2, Yuming Zhang1,2, Fengsong Liu1,2( ), Zhihua Ni1,2(
), Zhihua Ni1,2( )
)
- 1. School of Life Sciences, Hebei University, Baoding 071002, China
2. Key Laboratory of Zoological Systematics and Application of Hebei Province, Baoding 071002, China
-
Received:2023-02-10Revised:2023-05-01Online:2023-07-20Published:2023-05-15 -
Contact:Fengsong Liu,Zhihua Ni E-mail:huxiaoerfei@outlook.com;liufengsong@hbu.edu.cn;nizhihua@hbu.edu.cn -
Supported by:National Natural Science Foundation of China(42207336);Hebei Province Natural Science Foundation Joint Fund for Bioagriculture Project(C202204227);Post-graduates Innovation Fund Project of Hebei University(HBU2022ss016)
Cite this article
Feifei Li, Yun Wang, Jihai Gu, Yuming Zhang, Fengsong Liu, Zhihua Ni. E2F family play important roles in tumorigenesis[J]. Hereditas(Beijing), 2023, 45(7): 580-592.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2021, 71(3): 209-249.
doi: 10.3322/caac.v71.3 |
| [2] |
Siegel RL, Miller KD, Fuchs HE, Jemal A.Cancer statistics, 2022. CA Cancer J Clin, 2022, 72(1): 7-33.
doi: 10.3322/caac.v72.1 |
| [3] |
Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J Cell Sci, 2000, 113(Pt 10): 1661-1670.
doi: 10.1242/jcs.113.10.1661 |
| [4] |
Van Nostrand JL, Bowen ME, Vogel H, Barna M, Attardi LD. The p53 family members have distinct roles during mammalian embryonic development. Cell Death Differ, 2017, 24(4): 575-579.
doi: 10.1038/cdd.2016.128 pmid: 28211873 |
| [5] | Zhou DB, Xia Z, Xie MX, Gao Y, Yu Q, He BM.Exosomal long non-coding RNA SOX2 overlapping transcript enhances the resistance to EGFR-TKIs in non-small cell lung cancer cell line H1975. Hum Cell, 2021, 34(5): 1478-1489. |
| [6] |
Nacarino-Palma A, Rejano-Gordillo CM, González-Rico FJ, Ordiales-Talavero A, Román ÁC, Cuadrado M, Bustelo XR, Merino JM, Fernández-Salguero PM. Loss of aryl hydrocarbon receptor favors K-RasG12D-driven non- small cell lung cancer. Cancers (Basel), 2021, 13(16): 4071.
doi: 10.3390/cancers13164071 |
| [7] | Lei CG, Jia XY, Sun WJ. Establish six-gene prognostic model for glioblastoma based on multi-omics data of TCGA database. Hereditas (Beijing), 2021, 43(7): 665-689. |
| 雷常贵, 贾学渊, 孙文靖. 基于癌症基因组图谱计划多组学数据构建胶质母细胞瘤六基因预后模型. 遗传, 2021, 43(7): 665-689. | |
| [8] | Paul D. The systemic hallmarks of cancer. J Cancer Metastasis Treat, 2020, 6(8): 69-99. |
| [9] |
Jiramongkol Y, Lam EW. FOXO transcription factor family in cancer and metastasis. Cancer Metastasis Rev, 2020, 39(3): 681-709.
doi: 10.1007/s10555-020-09883-w |
| [10] | Kuo MH, Lee AC, Hsiao SH, Lin SE, Chiu YF, Yang LH, Yu CC, Chiou SH, Huang HN, Ko JC, Chou YT. Cross-talk between SOX2 and TGFβ signaling regulates EGFR-TKI tolerance and lung cancer dissemination. Cancer Res, 2020, 80(20): 4426-4438. |
| [11] |
Lee CS, Siprashvili Z, Mah A, Bencomo T, Elcavage LE, Che YL, Shenoy RM, Aasi SZ, Khavari PA. Mutant collagen COL11A1 enhances cancerous invasion. Oncogene, 2021, 40(44): 6299-6307.
doi: 10.1038/s41388-021-02013-y |
| [12] |
Bado IL, Zhang WJ, Hu JY, Xu Z, Wang H, Sarkar P, Li LC, Wan YW, Liu J, Wu W, Lo HC, Kim IS, Singh S, Janghorban M, Muscarella AM, Goldstein A, Singh P, Jeong HH, Liu CZ, Schiff R, Huang SX, Ellis MJ, Gaber MW, Gugala Z, Liu ZD, Zhang XHF. The bone microenvironment increases phenotypic plasticity of ER+ breast cancer cells. Dev Cell, 2021, 56(8): 1100-1117.
doi: 10.1016/j.devcel.2021.03.008 |
| [13] |
Park JH, Pyun WY, Park HW. Cancer metabolism: phenotype, signaling and therapeutic targets. Cells, 2020, 9(10): 2308.
doi: 10.3390/cells9102308 |
| [14] |
Masciale V, Grisendi G, Banchelli F, D'Amico R, Maiorana A, Sighinolfi P, Brugioni L, Stefani A, Morandi U, Dominici M, Aramini B. Cancer stem-like cells in a case of an inflammatory myofibroblastic tumor of the lung. Front Oncol, 2020, 10: 673.
doi: 10.3389/fonc.2020.00673 pmid: 32500024 |
| [15] |
Bodor JN, Boumber Y, Borghaei H. Biomarkers for immune checkpoint inhibition in non-small cell lung cancer (NSCLC). Cancer, 2020, 126(2): 260-270.
doi: 10.1002/cncr.32468 pmid: 31691957 |
| [16] |
Chen HH, Tarn WY. uORF-mediated translational control: recently elucidated mechanisms and implications in cancer. RNA Biol, 2019, 16(10): 1327-1338.
doi: 10.1080/15476286.2019.1632634 |
| [17] |
Yordy JS, Muise-Helmericks RC. Signal transduction and the ETS family of transcription factors. Oncogene, 2000, 19(55): 6503-6513.
pmid: 11175366 |
| [18] |
He XW, Lindsay-Mosher N, Li Y, Molinaro AM, Pellettieri J, Pearson BJ. FOX and ETS family transcription factors regulate the pigment cell lineage in planarians. Development, 2017, 144(24): 4540-4551.
doi: 10.1242/dev.156349 pmid: 29158443 |
| [19] |
Rajagopal C, Lankadasari MB, Aranjani JM, Harikumar KB. Targeting oncogenic transcription factors by polyphenols: a novel approach for cancer therapy. Pharmacol Res, 2018, 130: 273-291.
doi: S1043-6618(17)30968-4 pmid: 29305909 |
| [20] |
Tsigelny IF, Kouznetsova VL, Pingle SC, Kesari S. bHLH transcription factors inhibitors for cancer therapy: general features for in silico drug design. Curr Med Chem, 2014, 21(28): 3227-3243.
pmid: 24735358 |
| [21] |
Yeh JE, Toniolo PA, Frank DA. Targeting transcription factors: promising new strategies for cancer therapy. Curr Opin Oncol, 2013, 25(6): 652-658.
doi: 10.1097/01.cco.0000432528.88101.1a pmid: 24048019 |
| [22] |
Redell MS, Tweardy DJ. Targeting transcription factors for cancer therapy. Curr Pharm Des, 2005, 11(22): 2873-2887.
doi: 10.2174/1381612054546699 |
| [23] |
Oikawa T. ETS transcription factors: possible targets for cancer therapy. Cancer Sci, 2004, 95(8): 626-633.
doi: 10.1111/cas.2004.95.issue-8 |
| [24] |
Darnell JE Jr. Transcription factors as targets for cancer therapy. Nat Rev Cancer, 2002, 2(10): 740-749.
doi: 10.1038/nrc906 pmid: 12360277 |
| [25] | Mitra P. Targeting transcription factors in cancer drug discovery. Explor Target Antitumor Ther, 2020, 1(6): 401-412. |
| [26] |
Zhang WW, Yang SF, Chen DT, Yuwen DL, Zhang J, Wei XW, Han X, Guan XX. SOX2-OT induced by PAI-1 promotes triple-negative breast cancer cells metastasis by sponging miR-942-5p and activating PI3K/Akt signaling. Cell Mol Life Sci, 2022, 79(1): 59.
doi: 10.1007/s00018-021-04120-1 pmid: 34997317 |
| [27] |
Wei YY, Hou J, Tang WR, Luo Y. The cooperation between p53 and Ras in tumorigenesis. Hereditas (Beijing), 2012, 34(12): 1513-1521.
doi: 10.3724/SP.J.1005.2012.01513 |
|
魏永永, 侯静, 唐文如, 罗瑛. p53与Ras协同及其在肿瘤发生中的作用. 遗传, 2012, 34(12): 1513-1521.
doi: 10.3724/SP.J.1005.2012.01513 |
|
| [28] |
Bushweller JH. Targeting transcription factors in cancer- from undruggable to reality. Nat Rev Cancer, 2019, 19(11): 611-624.
doi: 10.1038/s41568-019-0196-7 pmid: 31511663 |
| [29] |
Lambert SA, Jolma A, Campitelli LF, Das PK, Yin YM, Albu M, Chen XT, Taipale J, Hughes TR, Weirauch MT. The human transcription factors. Cell, 2018, 172(4): 650-665.
doi: S0092-8674(18)30106-5 pmid: 29425488 |
| [30] |
Lambert M, Jambon S, Depauw S, David-Cordonnier MH. Targeting transcription factors for cancer treatment. Molecules, 2018, 23(6): 1479.
doi: 10.3390/molecules23061479 |
| [31] |
Xanthoulis A, Tiniakos DG. E2F transcription factors and digestive system malignancies: how much do we know? World J Gastroenterol, 2013, 19(21): 3189-3198.
doi: 10.3748/wjg.v19.i21.3189 |
| [32] |
Fischer M, Schade AE, Branigan TB, Müller GA, DeCaprio JA. Coordinating gene expression during the cell cycle. Trends Biochem Sci, 2022, 47(12): 1009-1022.
doi: 10.1016/j.tibs.2022.06.007 pmid: 35835684 |
| [33] |
Müller H, Helin K. The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta, 2000, 1470(1): M1-12.
doi: 10.1016/s0304-419x(99)00030-x pmid: 10656985 |
| [34] |
Wang QX, Liu JP, Cheang I, Li JH, Chen TZ, Li YX, Yu B. Comprehensive analysis of the E2F transcription factor family in human lung adenocarcinoma. Int J Gen Med, 2022, 15: 5973-5984.
doi: 10.2147/IJGM.S369582 pmid: 35811776 |
| [35] | Sun CC, Zhou Q, Hu W, Li SJ, Zhang F, Chen ZL, Li G, Bi ZY, Bi YY, Gong FY, Bo T, Yuan ZP, Hu WD, Zhan BT, Zhang Q, Tang QZ, Li DJ. Transcriptional E2F1/2/5/8 as potential targets and transcriptional E2F3/6/7 as new biomarkers for the prognosis of human lung carcinoma. Aging (Albany NY), 2018, 10(5): 973-987. |
| [36] |
Wang YY, Li M, Zhang L, Chen YT, Zhang SD. m6A demethylase FTO induces NELL2 expression by inhibiting E2F1 m6A modification leading to metastasis of non- small cell lung cancer. Mol Ther Oncolytics, 2021, 21: 367-376.
doi: 10.1016/j.omto.2021.04.011 |
| [37] |
El-Khattouti A, Sheehan NT, Monico J, Drummond HA, Haikel Y, Brodell RT, Megahed M, Hassan M. Cd133(+) melanoma subpopulation acquired resistance to caffeic acid phenethyl ester-induced apoptosis is attributed to the elevated expression of abcb5: significance for melanoma treatment. Cancer Lett, 2015, 357(1): 83-104.
doi: S0304-3835(14)00654-5 pmid: 25449786 |
| [38] |
Carvajal LA, Hamard PJ, Tonnessen C, Manfredi JJ. E2F7, a novel target, is up-regulated by p53 and mediates DNA damage-dependent transcriptional repression. Genes Dev, 2012, 26(14): 1533-1545.
doi: 10.1101/gad.184911.111 |
| [39] |
Li JX, Wang H, Cao FL, Cheng YF. A bioinformatics analysis for diagnostic roles of the E2F family in esophageal cancer. J Gastrointest Oncol, 2022, 13(5): 2115-2131.
doi: 10.21037/jgo |
| [40] |
Toolabi N, Daliri FS, Mokhlesi A, Talkhabi M. Identification of key regulators associated with colon cancer prognosis and pathogenesis. J Cell Commun Signal, 2022, 16(1): 115-127.
doi: 10.1007/s12079-021-00612-8 |
| [41] |
Liu XS, Gao Y, Liu C, Chen XQ, Zhou LM, Yang JW, Kui XY, Pei ZJ. Comprehensive analysis of prognostic and immune infiltrates for E2F transcription factors in human pancreatic adenocarcinoma. Front Oncol, 2020, 10: 606735.
doi: 10.3389/fonc.2020.606735 |
| [42] |
Shen C, Chen XM, Xiao K, Che GW. New relationship of E2F1 and BNIP3 with caveolin-1 in lung cancer- associated fibroblasts. Thorac Cancer, 2020, 11(6): 1369-1371.
doi: 10.1111/tca.v11.6 |
| [43] |
Xu ZH, Qu H, Ren YY, Gong ZZ, Ri HJ, Zhang F, Shao S, Chen XL, Chen X. Systematic analysis of E2F expression and its relation in colorectal cancer prognosis. Int J Gen Med, 2022, 15: 4849-4870.
doi: 10.2147/IJGM.S352141 pmid: 35585998 |
| [44] |
Weng JZ, Wu AX, Ying JW. Chemosensitivity of gastric cancer: analysis of key pathogenic transcription factors. J Gastrointest Oncol, 2022, 13(3): 977-984.
doi: 10.21037/jgo-22-274 pmid: 35837191 |
| [45] |
Hu H, Miao YR, Jia LH, Yu QY, Zhang Q, Guo AY. Animaltfdb 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res, 2019, 47(D1): D33-D38.
doi: 10.1093/nar/gky822 |
| [46] |
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 2010, 26(1): 139-140.
doi: 10.1093/bioinformatics/btp616 pmid: 19910308 |
| [47] |
Yu GC, Wang LG, Han YY, He QY. Clusterprofiler: an R package for comparing biological themes among gene clusters. Omics, 2012, 16(5): 284-287.
doi: 10.1089/omi.2011.0118 pmid: 22455463 |
| [48] |
Mendes FK, Vanderpool D, Fulton B, Hahn MW. CAFÉ5 models variation in evolutionary rates among gene families. Bioinformatics, 2020, 36(22-23): 5516-5518.
doi: 10.1093/bioinformatics/btaa1022 pmid: 33325502 |
| [49] |
Gaubatz S, Lindeman GJ, Ishida S, Jakoi L, Nevins JR, Livingston DM, Rempel RE. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell, 2000, 6(3): 729-735.
pmid: 11030352 |
| [50] |
Shiah JV, Johnson DE, Grandis JR. Transcription factors and cancer: approaches to targeting. Cancer J, 2023, 29(1): 38-46.
doi: 10.1097/PPO.0000000000000639 pmid: 36693157 |
| [51] | Shen C, Li J, Chang S, Che GW. Advancement of E2F1 in common tumors. Chin J Lung Cancer, 2020, 23(10): 921-926. |
| 沈诚, 李珏, 常帅, 车国卫. E2F1在常见肿瘤中的最新研究进展. 中国肺癌杂志, 2020, 23(10): 921-926. | |
| [52] |
Cam H, Dynlacht BD. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell, 2003, 3(4): 311-316.
doi: 10.1016/s1535-6108(03)00080-1 pmid: 12726857 |
| [53] |
Xie D, Pei Q, Li JY, Wan X, Ye T. Emerging role of E2F family in cancer stem cells. Front Oncol, 2021, 11: 723137.
doi: 10.3389/fonc.2021.723137 |
| [54] |
Jin X, Ding DL, Yan YQ, Li H, Wang B, Ma LL, Ye ZQ, Ma T, Wu Q, Rodrigues DN, Kohli M, Jimenez R, Wang LG, Goodrich DW, de Bono J, Dong HD, Wu HS, Zhu RZ, Huang HJ. Phosphorylated RB promotes cancer immunity by inhibiting NF-κb activation and PD-L1 expression. Mol Cell, 2019, 73(1): 22-35.e26.
doi: S1097-2765(18)30894-3 pmid: 30527665 |
| [55] |
Gulluni F, Prever L, Li HY, Krafcikova P, Corrado I, Lo WT, Margaria JP, Chen A, De Santis MC, Cnudde SJ, Fogerty J, Yuan A, Massarotti A, Sarijalo NT, Vadas O, Williams RL, Thelen M, Powell DR, Schueler M, Wiesener MS, Balla T, Baris HN, Tiosano D, McDermott BM Jr, Perkins BD, Ghigo A, Martini M, Haucke V, Boura E, Merlo GR, Buchner DA, Hirsch E. PI(3,4)P2- mediated cytokinetic abscission prevents early senescence and cataract formation. Science, 2021, 374(6573): eabk0410.
doi: 10.1126/science.abk0410 |
| [56] | Segeren HA, Westendorp B. Mechanisms used by cancer cells to tolerate drug-induced replication stress. Cancer Lett, 2022, 544: 215804. |
| [57] | Guo AY, Hu JL, Li YY, Li P. Association of a low-frequency missense variant in E2F transcription factor 7 with colorectal cancer risk. Chin J Health Lab Technol, 2018, 28(17): 2118-2122. |
| 郭爱叶, 胡金龙, 李颖颖, 李泮. E2F转录因子7低频错义突变与结直肠癌风险的相关性研究. 中国卫生检验杂志, 2018, 28(17): 2118-2122. |
| [1] | Kai Chen, Hao Wang, Yiting Chen, Ke Fu, Zhigang Han, Cong Li, Jinping Si, Donghong Chen. Functional analysis of WOX family genes in Dendrobium catenatum during growth and development [J]. Hereditas(Beijing), 2023, 45(8): 700-714. |
| [2] | Shuang Zhang, Shanshan Guo, Ruwen Wang, Renyan Ma, Xianmin Wu, Peijie Chen, Ru Wang. The roles of PARK gene family in myopathy [J]. Hereditas(Beijing), 2022, 44(7): 545-555. |
| [3] | Xiaocui Li, Kaicheng Kang, Xianzhong Huang, Yongbin Fan, Miaomiao Song, Yunjie Huang, Jiajia Ding. Genome-wide identification, phylogenetic analysis and expression profiling of the MKK gene family in Arabidopsis pumila [J]. Hereditas(Beijing), 2020, 42(4): 403-421. |
| [4] | Taotao Wang, Yong Yang, Wei Wei, Chentao Lin, Liuyin Ma. Identification and expression analyses of the NAC transcription factor family in Spartina alterniflora [J]. Hereditas(Beijing), 2020, 42(2): 194-211. |
| [5] | Yu Meng,Ruolin Yang. Comparative analysis of gene family size provides insight into the adaptive evolution of vertebrates [J]. Hereditas(Beijing), 2019, 41(2): 158-174. |
| [6] | Ming Li,Feiyue Cheng,Luyao Gong,Hua Xiang. Systematic discovery of novel prokaryotic defense systems: progress and prospects [J]. Hereditas(Beijing), 2018, 40(4): 259-265. |
| [7] | Zongchang Xu,Yingzhen Kong. Genome-wide identification, subcellular localization and gene expression analysis of the members of CESA gene family in common tobacco (Nicotiana tabacum L.) [J]. Hereditas(Beijing), 2017, 39(6): 512-524. |
| [8] | Shuaiqi Zhu,Yifu Gong,Li Zhang,Kai Yu,Heyu Wang,Xiaojun Yan. Expression analysis of different β-carotenoid hydroxylase gene families in stress response in Dunaliella viridis [J]. Hereditas(Beijing), 2017, 39(2): 156-165. |
| [9] | Yimin He, Mingmin Gu. Research progress of myosin heavy chain genes in human genetic diseases [J]. Hereditas(Beijing), 2017, 39(10): 877-887. |
| [10] | Xiaohua Xiang, Xinru Wu, Jiangtao Chao, Minglei Yang, Fan Yang, Guo Chen, Guanshan Liu, Yuanying Wang. Genome-wide identification and expression analysis of the WRKY gene family in common tobacco (Nicotiana tabacum L.) [J]. Hereditas(Beijing), 2016, 38(9): 840-856. |
| [11] | Jianzhong Chang, Fengxia Yan, Linyi Qiao, Jun Zheng, Fuyao Zhang, Qingshan Liu. Genome-wide identification and expression analysis of SBP-box gene family in Sorghum bicolor L. [J]. HEREDITAS(Beijing), 2016, 38(6): 569-580. |
| [12] | Minglei Yang, Jiangtao Chao, Dawei Wang, Junhua Hu, Hua Wu, Daping Gong, Guanshan Liu. Genome-wide identification and expression profiling of the C2H2-type zinc finger protein transcription factor family in tobacco [J]. HEREDITAS(Beijing), 2016, 38(4): 337-349. |
| [13] | Yanbing Gu, Zhirui Ji, Fumei Chi, Zhuang Qiao, Chengnan Xu, Junxiang Zhang, Zongshan Zhou, Qinglong Dong. Genome-wide identification and expression analysis of the WRKY gene family in peach [J]. HEREDITAS(Beijing), 2016, 38(3): 254-270. |
| [14] | Huazhao Yuan, Mizhen Zhao, Weimin Wu, HongMei Yu, Yaming Qian, Zhuangwei Wang, Xicheng Wang. Genome-wide identification and expression analysis of auxin-related gene families in grape [J]. HEREDITAS(Beijing), 2015, 37(7): 720-730. |
| [15] | Yang Shi, Xiao Xu, Haoyang Li, Qian Xu, Jichen Xu. Bioinformatics analysis of the expansin gene family in rice [J]. HEREDITAS(Beijing), 2014, 36(8): 809-820. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||