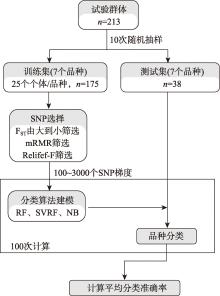
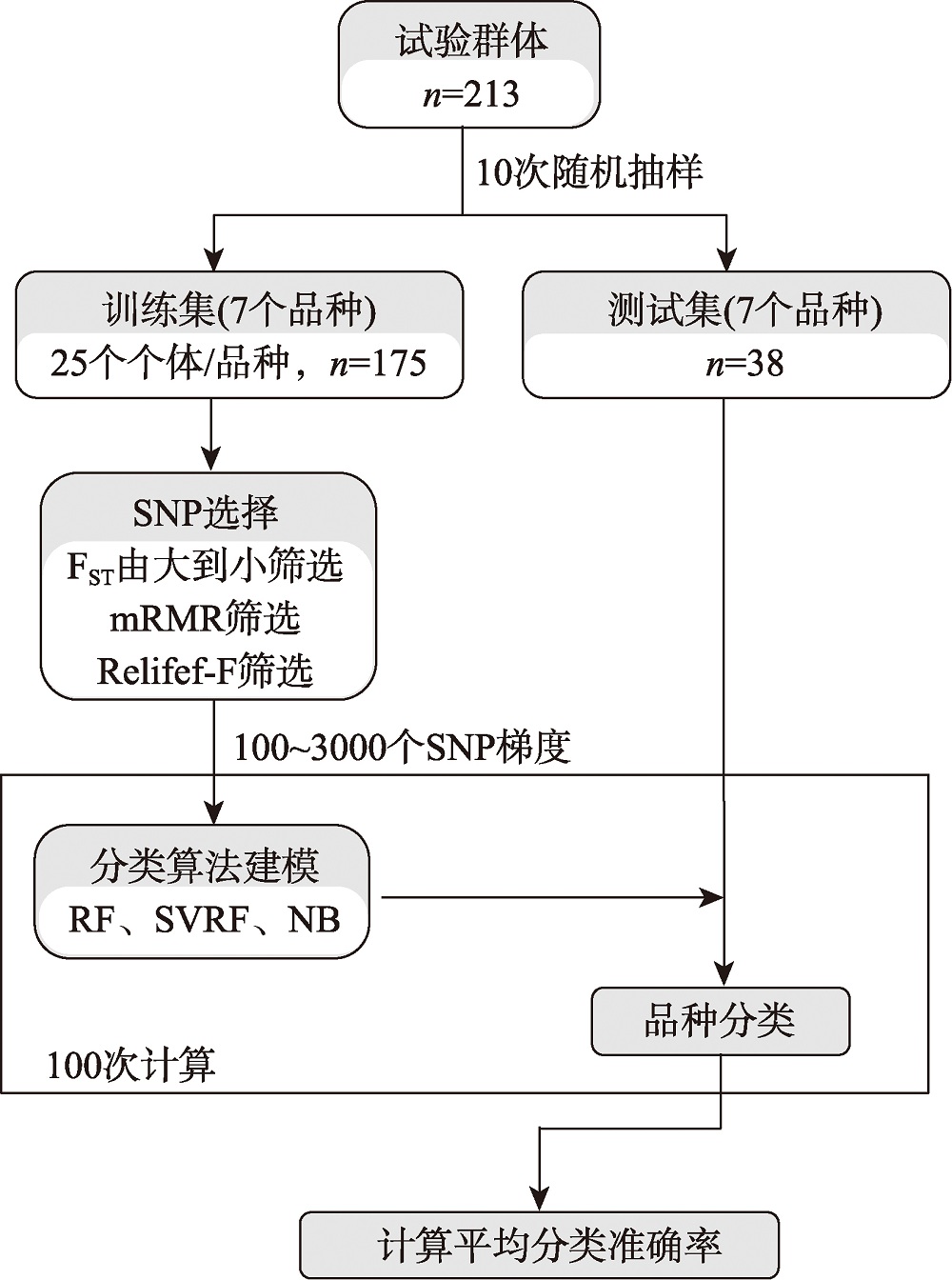
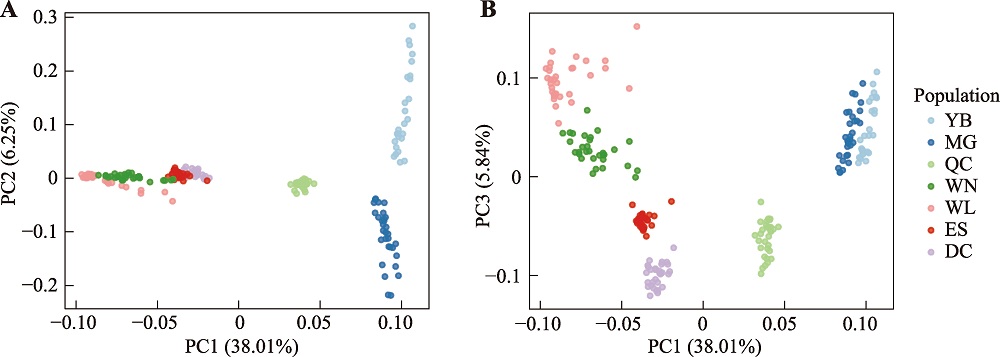
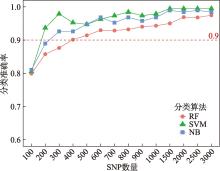
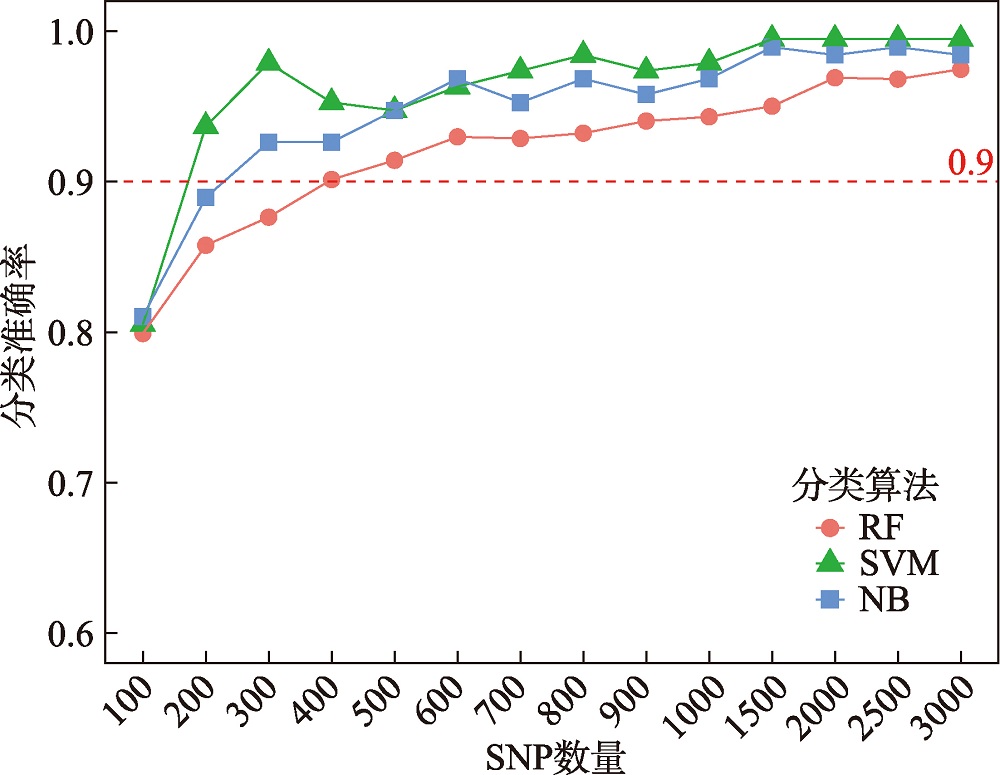
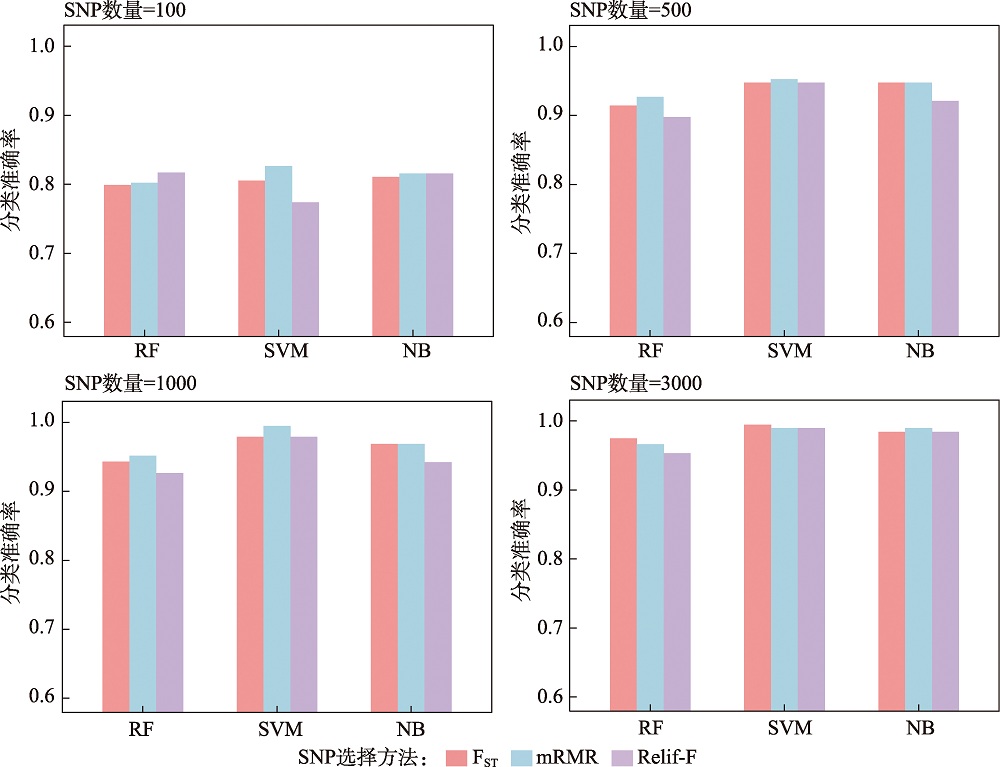
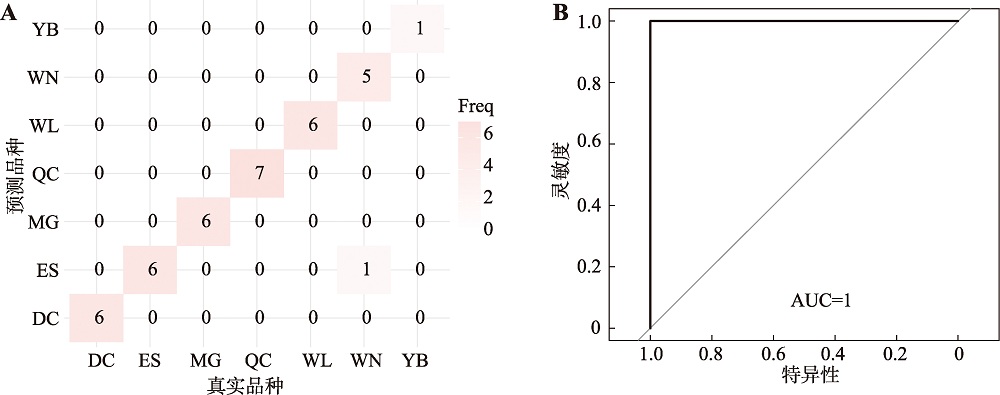
Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (7): 530-539.doi: 10.16288/j.yczz.24-059
• Research Article • Previous Articles Next Articles
Classification accuracy of machine learning algorithms for Chinese local cattle breeds using genomic markers
Hui Liang( ), Xue Wang, Jingfang Si, Yi Zhang(
), Xue Wang, Jingfang Si, Yi Zhang( )
)
- College of Animal Science and Technology, China Agricultural University, Beijing 100193, China
-
Received:2024-03-11Revised:2024-06-25Online:2024-07-20Published:2024-06-26 -
Supported by:National Key Research and Development Program of China(2021YFD1200904);Earmarked Fund for CARS36(CARS-36)
Cite this article
Hui Liang, Xue Wang, Jingfang Si, Yi Zhang. Classification accuracy of machine learning algorithms for Chinese local cattle breeds using genomic markers[J]. Hereditas(Beijing), 2024, 46(7): 530-539.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Sun H, Olasege BS, Xu Z, Zhao QB, Ma PP, Wang QS, Lu SX, Pan YC. Genome-wide and trait-specific markers: a perspective in designing conservation programs. Front Genet, 2018, 9: 389.
doi: 10.3389/fgene.2018.00389 pmid: 30283493 |
| [2] |
Maudet C, Luikart G, Taberlet P. Genetic diversity and assignment tests among seven French cattle breeds based on microsatellite DNA analysis. J Anim Sci, 2002, 80(4): 942-950.
pmid: 12002331 |
| [3] |
Suekawa Y, Aihara H, Araki M, Hosokawa D, Mannen H, Sasazaki S. Development of breed identification markers based on a bovine 50K SNP array. Meat Sci, 2010, 85(2): 285-288.
doi: 10.1016/j.meatsci.2010.01.015 pmid: 20374900 |
| [4] | Lewis J, Abas Z, Dadousis C, Lykidis D, Paschou P, Drineas P. Tracing cattle breeds with principal components analysis ancestry informative SNPs. PLoS One, 2011, 6(4): e18007. |
| [5] |
Putnová L, Štohl R. Comparing assignment-based approaches to breed identification within a large set of horses. J Appl Genet, 2019, 60(2): 187-198.
doi: 10.1007/s13353-019-00495-x pmid: 30963515 |
| [6] | Gao J, Sun LW, Zhang SS, Xu JH, He MQ, Zhang DF, Wu CF, Dai JJ. Screening discriminating SNPs for Chinese indigenous pig breeds identification using a random forests algorithm. Genes (Basel), 2022, 13(12): 2207. |
| [7] |
Sharma A, Dey P. A machine learning approach to unmask novel gene signatures and prediction of Alzheimer's disease within different brain regions. Genomics, 2021, 113(4): 1778-1789.
doi: 10.1016/j.ygeno.2021.04.028 pmid: 33878365 |
| [8] |
Zhang ZS, Liu ZP. Robust biomarker discovery for hepatocellular carcinoma from high-throughput data by multiple feature selection methods. BMC Med Genomics, 2021, 14(Suppl 1): 112.
doi: 10.1186/s12920-021-00957-4 pmid: 34433487 |
| [9] | Yang YL, Wang XY, Wang SY, Chen Q, Li M L, Lu SX. Identification of potential sex-specific biomarkers in pigs with low and high intramuscular fat content using integrated bioinformatics and machine learning. Genes (Basel), 2023, 14(9): 1695. |
| [10] |
Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution, 1984, 38(6): 1358-1370.
doi: 10.1111/j.1558-5646.1984.tb05657.x pmid: 28563791 |
| [11] |
Schiavo G, Bertolini F, Galimberti G, Bovo S, Dall’Olio S, Nanni Costa L, Gallo M, Fontanesi L. A machine learning approach for the identification of population-informative markers from high-throughput genotyping data: application to several pig breeds. Animal, 2020, 14(2): 223-232.
doi: 10.1017/S1751731119002167 pmid: 31603060 |
| [12] | Zhao CH, Wang D, Teng J, Yang C, Zhang XY, Wei XM, Zhang Q. Breed identification using breed-informative SNPs and machine learning based on whole genome sequence data and SNP chip data. J Anim Sci Biotechnol, 2023, 14(1): 85. |
| [13] | Kumar H, Panigrahi M, Chhotaray S, Parida S, Chauhan A, Bhushan B, Gaur GK, Mishra BP, Singh RK. Comparative analysis of five different methods to design a breed- specific SNP panel for cattle. Anim Biotechnol, 2021, 32(1): 130-136. |
| [14] | Mahendran N, Durai Raj Vincent PM, Srinivasan K, Chang CY. Machine learning based computational gene selection models: a survey, performance evaluation, open issues, and future research directions. Front Genet, 2020, 11: 603808 |
| [15] | Liu RQ, Xu ZT, Teng JY, Pan XC, Lin Q, Cai XD, Diao SQ, Feng XY, Yuan XL, Li JQ, Zhang Z. Evaluation of six machine learning classification algorithms in pig breed identification using SNPs array data. Anim Genet, 2023, 54(2): 113-122. |
| [16] |
Pasupa K, Rathasamuth W, Tongsima S. Discovery of significant porcine SNPs for swine breed identification by a hybrid of information gain, genetic algorithm, and frequency feature selection technique. BMC Bioinformatics, 2020, 21(1): 216.
doi: 10.1186/s12859-020-3471-4 pmid: 32456608 |
| [17] |
Hayah I, Ababou M, Botti S, Badaoui B. Comparison of three statistical approaches for feature selection for fine-scale genetic population assignment in four pig breeds. Trop Anim Health Prod, 2021, 53(3): 395.
doi: 10.1007/s11250-021-02824-x pmid: 34245361 |
| [18] |
Hulsegge B, Calus MPL, Windig JJ, Hoving-Bolink AH, Maurice-van Eijndhoven MHT, Hiemstra SJ. Selection of SNP from 50K and 777K arrays to predict breed of origin in cattle. J Anim Sci, 2013, 91(11): 5128-5134.
doi: 10.2527/jas.2013-6678 pmid: 24045484 |
| [19] |
Judge MM, Kelleher MM, Kearney JF, Sleator RD, Berry DP. Ultra-low-density genotype panels for breed assignment of Angus and Hereford cattle. Animal, 2017, 11(6): 938-947.
doi: 10.1017/S1751731116002457 pmid: 27881206 |
| [20] |
Liu YX, Zhang NN, He Y, Lun LJ. Prediction of core cancer genes using a hybrid of feature selection and machine learning methods. Genet Mol Res, 2015, 14(3): 8871-8882.
doi: 10.4238/2015.August.3.10 pmid: 26345818 |
| [21] | Shreem SS, Abdullah S, Nazri MZA, Alzaqebah MA. Hybridizing ReliefF, MRMR filters and GA wrapper approaches for gene selection. J Theor Appl Inf Technol, 2012, 46(2): 1034-1039. |
| [22] |
Gao YH, Gautier M, Ding XD, Zhang H, Wang YC, Wang X, Faruque MO, Li JY, Ye SH, Gou X, Han JL, Lenstra JA, Zhang Y. Species composition and environmental adaptation of indigenous Chinese cattle. Sci Rep, 2017, 7(1): 16196.
doi: 10.1038/s41598-017-16438-7 pmid: 29170422 |
| [23] |
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet, 2007, 81(3): 559-575.
doi: 10.1086/519795 pmid: 17701901 |
| [24] | Wickham H. ggplot2: elegant graphics for data analysis. Springer New York, 2009. |
| [25] | Hanchuan P, Fuhui L, Chris D. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell, 2005, 27(8): 1226-1238. |
| [26] |
De Jay N, Papillon-Cavanagh S, Olsen C, El-Hachem N, Bontempi G, Haibe-Kains B. mRMRe: an R package for parallelized mRMR ensemble feature selection. Bioinformatics, 2013, 29(18): 2365-2368.
doi: 10.1093/bioinformatics/btt383 pmid: 23825369 |
| [27] | Robnik-Šikonja M, Kononenko I. Theoretical and empirical analysis of ReliefF and RReliefF. Mach Learn, 2003, 53(1): 23-69. |
| [28] | Robnik-Šikonja M, Savicky P. CORElearn: classification, regression and feature evaluation. 2020. |
| [29] | Liaw A, Wiener M. Classification and regression by RandomForest. Forest, 2001, 23(2/3): 18-22. |
| [30] | Meyer D. Support Vector Machines∗ the interface to libsvm in package e1071. 2001. |
| [31] |
Zhang ZH. Naïve Bayes classification in R. Ann Transl Med, 2016, 4(12): 241.
doi: 10.21037/atm.2016.03.38 pmid: 27429967 |
| [32] | Xu ZT, Diao SQ, Teng JY, Chen ZT, Feng XY, Cai XT, Yuan XL, Zhang H, Li JQ, Zhang Z. Breed identification of meat using machine learning and breed tag SNPs. Food Control, 2021, 125: 107971. |
| [33] |
Bertolini F, Galimberti G, Calò DG, Schiavo G, Matassino D, Fontanesi L. Combined use of principal component analysis and random forests identify population- informative single nucleotide polymorphisms: application in cattle breeds. J Anim Breed Genet, 2015, 132(5): 346-356.
doi: 10.1111/jbg.12155 pmid: 25781205 |
| [34] |
Li B, Zhang NX, Wang YG, George AW, Reverter A, Li YT. Genomic prediction of breeding values using a subset of SNPs identified by three machine learning methods. Front Genet, 2018, 9: 237.
doi: 10.3389/fgene.2018.00237 pmid: 30023001 |
| [35] | Yang JF, Qiao PR, Li YM, Wang N. A review of machine-learning classification and algorithms. Statistics & Decision, 2019, 35(06): 36-40. |
| 杨剑锋, 乔佩蕊, 李永梅, 王宁. 机器学习分类问题及算法研究综述. 统计与决策, 2019, 35(6): 36-40. | |
| [36] | Zhang Y, Ding C, Li T. Gene selection algorithm by combining reliefF and mRMR. BMC Genomics, 2008, 9(2): S27. |
| [37] | Wilmot H, Bormann J, Soyeurt H, Hubin X, Glorieux G, Mayeres P, Bertozzi C, Gengler N. Development of a genomic tool for breed assignment by comparison of different classification models: application to three local cattle breeds. J Anim Breed Genet, 2022, 139(1): 40-61. |
| [38] | Huang JJ. Identify pig breeds with different methods based on SNP chip[Dissertation]. South China Agricultural University, 2019. |
| 黄进杰. 基于SNP芯片利用不同方法鉴定个体猪品种[学位论文]. 华南农业大学, 2019. |
| [1] | Dong Chen, Shujie Wang, Zhenjian Zhao, Xiang Ji, Qi Shen, Yang Yu, Shengdi Cui, Junge Wang, Ziyang Chen, Jinyong Wang, Zongyi Guo, Pingxian Wu, Guoqing Tang. Genomic prediction of pig growth traits based on machine learning [J]. Hereditas(Beijing), 2023, 45(10): 922-932. |
| [2] | Yongqiang Kong, Jinkai Liu, Jiaqi Gu, Jingyi Xu, Yunuo Zheng, Yiliang Wei, Shaoyuan Wu. Optimization scheme of machine learning model for genetic division between northern Han, southern Han, Korean and Japanese [J]. Hereditas(Beijing), 2022, 44(11): 1028-1043. |
| [3] | Yali Hu, Rui Dai, Yongxin Liu, Jingying Zhang, Bin Hu, Chengcai Chu, Huaibo Yuan, Yang Bai. Analysis of rice root bacterial microbiota of Nipponbare and IR24 [J]. Hereditas(Beijing), 2020, 42(5): 506-518. |
| [4] | Zhao Xuetong, Yang Yadong, Qu Hongzhu, Fang Xiangdong. Applications of machine learning in clinical decision support in the omic era [J]. Hereditas(Beijing), 2018, 40(9): 693-703. |
| [5] | Zhang Guishan, Yang Yong, Zhang Lingmin, Dai Xianhua. Application of machine learning in the CRISPR/Cas9 system [J]. Hereditas(Beijing), 2018, 40(9): 704-723. |
| [6] | Zhe-ye Peng,Zi-jun Tang,Min-zhu Xie. Research progress in machine learning methods for gene-gene interaction detection [J]. Hereditas(Beijing), 2018, 40(3): 218-226. |
| [7] | HOU Yan-Yan, YING Xiao-Min, LI Wu-Ju . Computational approaches to microRNA discovery [J]. HEREDITAS, 2008, 30(6): 687-696. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||