Hereditas(Beijing) ›› 2025, Vol. 47 ›› Issue (4): 437-447.doi: 10.16288/j.yczz.24-239
• Review • Previous Articles Next Articles
The role of cis-regulatory elements in the determination and transformation of muscle fiber type in animal skeletal muscles
Jing Luo1( ), Kaiying Lei2, Song Shi1, Xiaoli Xu1, Xueliang Sun1, Meijun Song1, Hongping Zhang1, Li Li1(
), Kaiying Lei2, Song Shi1, Xiaoli Xu1, Xueliang Sun1, Meijun Song1, Hongping Zhang1, Li Li1( )
)
- 1. College of Animal Science and Technology, Sichuan Agricultural University, Chengdu 611130, China
2. Aba Secondary Vocational and Technical Schools, Jiuzhaigou 623400, China
-
Received:2024-08-16Revised:2024-12-19Online:2025-04-20Published:2025-01-16 -
Contact:Li Li E-mail:luojing11032021@126.com;lily@sicau.edu.cn -
Supported by:National Natural Science Foundation of China(32072715)
Cite this article
Jing Luo, Kaiying Lei, Song Shi, Xiaoli Xu, Xueliang Sun, Meijun Song, Hongping Zhang, Li Li. The role of cis-regulatory elements in the determination and transformation of muscle fiber type in animal skeletal muscles[J]. Hereditas(Beijing), 2025, 47(4): 437-447.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Characteristics of four muscle fiber types"
| 肌纤维类型 | MyHC基因 | 单克隆抗体 | 功能特征 | 肉质特征 | 参考文献 |
|---|---|---|---|---|---|
| I | Myh7 | BAF8、NOQ7.5.4D、BA-D5、A4.840、4B51E8 | 收缩速度慢、力量小、恢复快、持续时间长 | 深红色、pH大、系水力强、剪切力小、肉质嫩、风味好 | [ |
| IIA | Myh2 | A4.74、SC-71、2F7、8F72C8 | 中等收缩速度、中等力量、持续时间较长 | 红色、pH较高、系水力较强、剪切力较小、肉质较嫩、风味较好 | [ |
| IIX | Myh1 | BF-35、6H1、6F12H3 | 收缩速度较快、力量较大、持续时间较短 | 浅红色、pH较低、系水力较弱、剪切力较大、肉质较粗糙、风味较差 | [ |
| IIB | Myh4 | BF-F3、10F5、2G72F10 | 收缩速度快、力量大、易疲劳、恢复慢 | 白色、pH低、系水力弱、剪切力大、肉质粗糙、风味差 | [ |
| [1] |
De Deyne PG, Hayatsu K, Meyer R, Paley D, Herzenberg JE. Muscle regeneration and fiber-type transformation during distraction osteogenesis. J Orthop Res, 1999, 17(4): 560-570.
pmid: 10459763 |
| [2] |
Klont RE, Brocks L, Eikelenboom G. Muscle fibre type and meat quality. Meat Sci, 1998, 49(S1): S219-S229.
pmid: 22060713 |
| [3] |
Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev, 2011, 91(4): 1447-1531.
pmid: 22013216 |
| [4] |
Kanning KC, Kaplan A, Henderson CE. Motor neuron diversity in development and disease. Annu Rev Neurosci, 2010, 33: 409-440.
pmid: 20367447 |
| [5] |
Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev, 1998, 12(16): 2499-2509.
pmid: 9716403 |
| [6] |
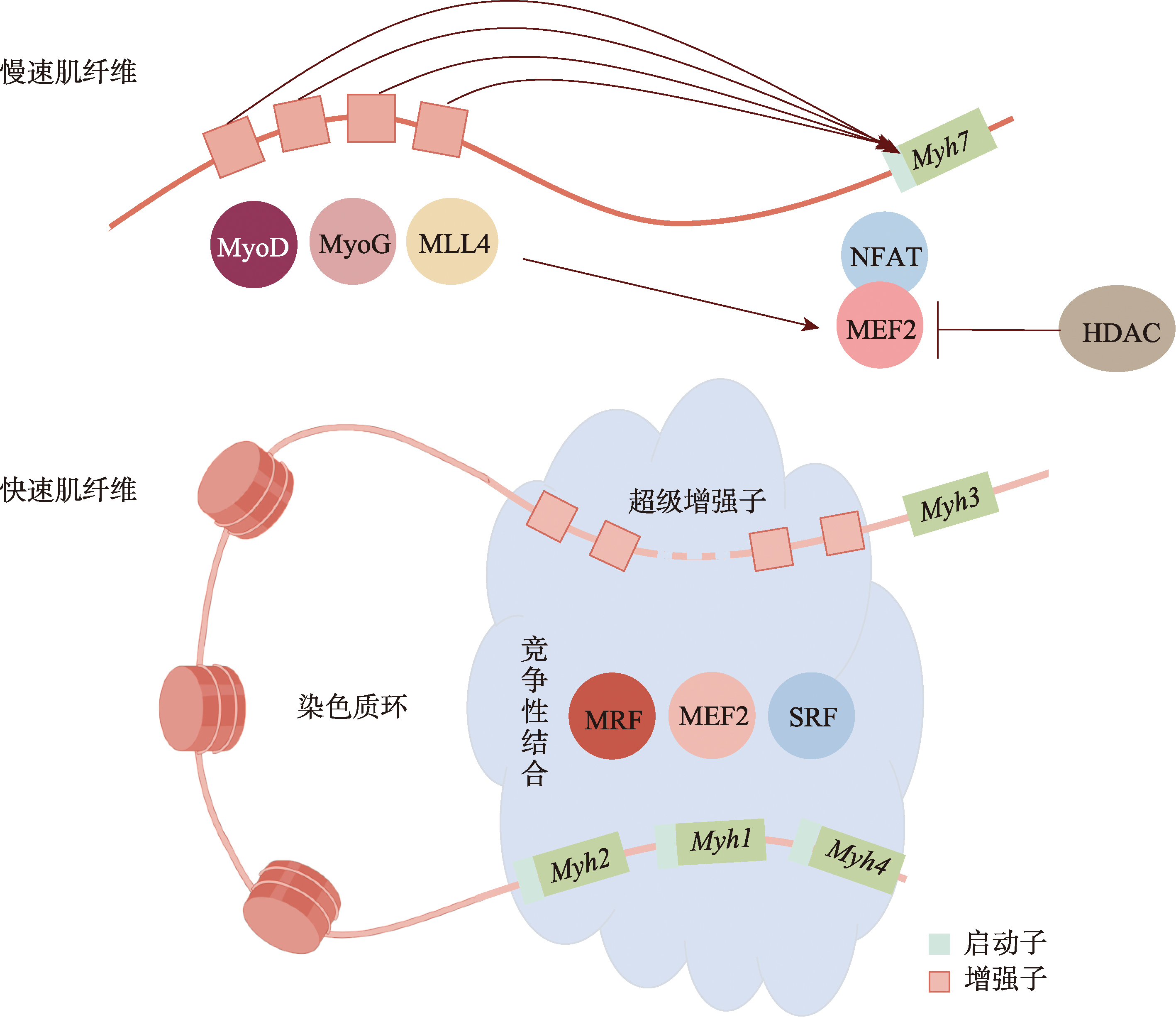
Dos Santos M, Backer S, Auradé F, Wong MM, Wurmser M, Pierre R, Langa F, Do CM, Schmitt A, Concordet JP, Sotiropoulos A, Jeffrey Dilwoeth F, Noordermeer D, Relaix F, Sakakibara I, Maire P. A fast myosin super enhancer dictates muscle fiber phenotype through competitive interactions with myosin genes. Nat Commun, 2022, 13(1): 1039.
pmid: 35210422 |
| [7] |
Larson L, Lioy J, Johnson J, Medler S. Transitional hybrid skeletal muscle fibers in rat soleus development. J Histochem Cytochem, 2019, 67(12): 891-900.
pmid: 31510854 |
| [8] |
Brooke MH, Kaiser KK. Three “myosin adenosine triphosphatase” systems: the nature of their phlability and sulfhydryl dependence. J Histochem Cytochem, 1970, 18(9): 670-672.
pmid: 4249441 |
| [9] |
Peter JB, Sawaki S, Barnard RJ, Edgerton VR, Gillespie CA. Lactate dehydrogenase isoenzymes: distribution in fast-twitch red, fast-twitch white, and slow-twitch intermediate fibers of guinea pig skeletal muscle. Arch Biochem Biophys, 1971, 144(1): 304-307.
pmid: 5117531 |
| [10] |
Close RI. Dynamic properties of mammalian skeletal muscles. Physiol Rev, 1972, 52(1): 129-197.
pmid: 4256989 |
| [11] |
Ashmore CR, Doerr L. Comparative aspects of muscle fiber types in different species. Exp Neurol, 1971, 31(3): 408-418.
pmid: 4254914 |
| [12] |
Peter JB, Barnard RJ, Edgerton VR, Gillespie CA, Stempel KE. Metabolic profiles of three fiber types of skeletal muscle in guinea pigs and rabbits. Biochemistry, 1972, 11(14): 2627-2633.
pmid: 4261555 |
| [13] | Chen L. Research progress on the classification and factors affecting of skeletal muscle fiber in livestock. Chinese Journal of Animal Science, 2016, 52(19): 99-103. |
| 陈利. 家畜骨骼肌纤维分类及影响因素的研究进展. 中国畜牧杂志, 2016, 52(19): 99-103. | |
| [14] | Liang TY, Wu JP, Liu T, Bai Y, Zhang R. Recent progress in classification and transformation mechanism of muscle fiber types. Meat Research, 2018, 32(9): 55-61. |
| 梁婷玉, 吴建平, 刘婷, 柏妍, 张瑞. 肌纤维类型分类及转化机理研究进展. 肉类研究, 2018, 32(9): 55-61. | |
| [15] |
Sawano S, Mizunoya W. History and development of staining methods for skeletal muscle fiber types. Histol Histopathol, 2022, 37(6): 493-503.
pmid: 35043970 |
| [16] |
Liu T, Bai YP, Wang CL, Zhang TW, Su RN, Wang BH, Duan Y, Sun LN, Jin Y, Su L. Effects of probiotics supplementation on the intestinal metabolites, muscle fiber properties, and meat quality of sunit lamb. Animals (Basel), 2023, 13(4): 762.
pmid: 36830552 |
| [17] |
Mo MJ, Zhang ZH, Wang XT, Shen WJ, Zhang L, Lin SD. Molecular mechanisms underlying the impact of muscle fiber types on meat quality in livestock and poultry. Front Vet Sci, 2023, 10: 1284551.
pmid: 38076559 |
| [18] |
Rossi AC, Mammucari C, Argentini C, Reggiani C, Schiaffino S. Two novel/ancient myosins in mammalian skeletal muscles: MYH14/7b and MYH15 are expressed in extraocular muscles and muscle spindles. J Physiol, 2010, 588(Pt 2): 353-364.
pmid: 19948655 |
| [19] |
Schiaffino S, Hughes SM, Murgia M, Reggiani C. Myh13, a superfast myosin expressed in extraocular, laryngeal and syringeal muscles. J Physiol, 2024, 602(3): 427-443.
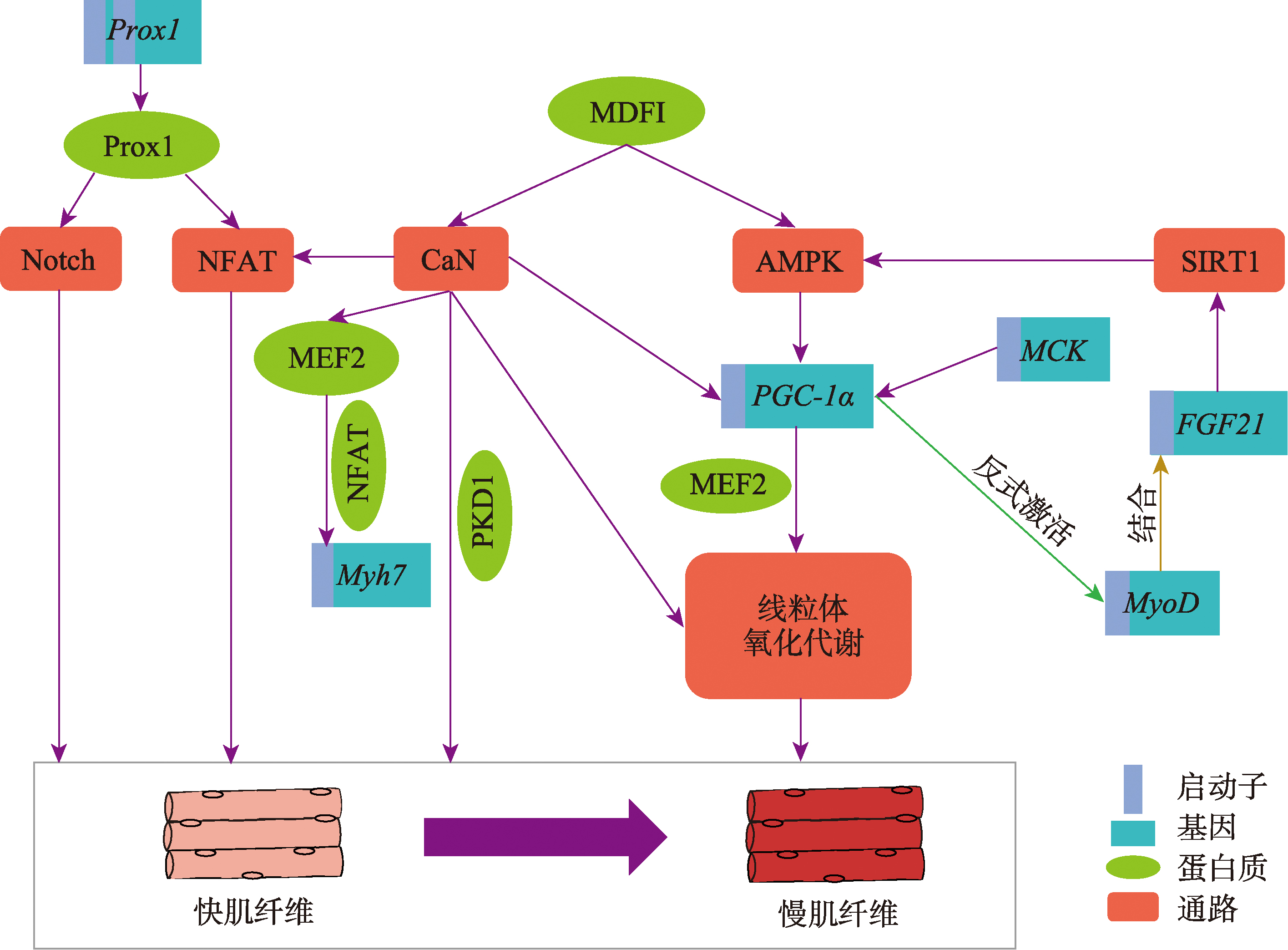
pmid: 38160435 |
| [20] |
Qin H, Hsu MKH, Morris BJ, Hoh JFY. A distinct subclass of mammalian striated myosins: structure and molecular evolution of "superfast" or masticatory myosin heavy chain. J Mol Evol, 2002, 55(5): 544-552.
pmid: 12399928 |
| [21] |
Hemmings KM, Parr T, Daniel ZCTR, Picard B, Buttery PJ, Brameld JM. Examination of myosin heavy chain isoform expression in ovine skeletal muscles. J Anim Sci, 2009, 87(12): 3915-3922.
pmid: 19684280 |
| [22] |
Argüello A, López-Fernández JL, Rivero JL. Limb myosin heavy chain isoproteins and muscle fiber types in the adult goat (Capra hircus). Anat Rec, 2001, 264(3): 284-293.
pmid: 11596010 |
| [23] | Guan SY, Gu XL, Shen HX, Ma YM, Chen L, Qu L, Qin LH. Characteristics and types of muscle fibers in different parts of Chinese red steppe cattle. Meat Research, 2023, 37(11): 8-12. |
| 关诗宇, 谷兴亮, 沈宏旭, 马玉敏, 程雷, 曲磊, 秦立红. 中国草原红牛不同部位肌纤维特性及类型. 肉类研究, 2023, 37(11): 8-12. | |
| [24] |
Deng KP, Liu ZP, Su YL, Fan YX, Zhang YL, Wang F. Comparison of muscle fiber characteristics and meat quality between newborn and adult haimen goats. Meat Sci, 2024, 207: 109361.
pmid: 37857027 |
| [25] |
Nuzzo JL. Sex differences in skeletal muscle fiber types: a meta-analysis. Clin Anat, 2024, 37(1): 81-91.
pmid: 37424380 |
| [26] |
Prasongsook S, Choi I, Bates RO, Raney NE, Ernst CW, Tumwasorn S. Association of insulin-like growth factor binding protein 2 genotypes with growth, carcass and meat quality traits in pigs. J Anim Sci Technol, 2015, 57: 31.
pmid: 26339502 |
| [27] |
Lu ZK, Yue YJ, Shi HN, Zhang JX, Liu TG, Liu JB, Yang BH. Effects of sheep sires on muscle fiber characteristics, fatty acid composition and volatile flavor compounds in F1 crossbred lambs. Foods, 2022, 11(24): 4076.
pmid: 36553818 |
| [28] |
Huo WR, Weng KQ, Gu TT, Zhang Y, Zhang Y, Chen GH, Xu Q. Effect of muscle fiber characteristics on meat quality in fast- and slow-growing ducks. Poult Sci, 2021, 100(8): 101264.
pmid: 34174572 |
| [29] |
Wu F, Zuo JJ, Yu QP, Zou SG, Tan HZ, Xiao J, Liu YH, Feng DY. Effect of skeletal muscle fibers on porcine meat quality at different stages of growth. Genet Mol Res, 2015, 14(3): 7873-7882.
pmid: 26214468 |
| [30] | Listrat A, Gagaoua M, Andueza D, Gruffat D, Normand J, Mairesse G, Picard B, Hocquette J. What are the drivers of beef sensory quality using metadata of intramuscular connective tissue, fatty acids and muscle fiber characteristics? Livest Sci, 2020, 240: 104209. |
| [31] |
Wang H, Fan Z, Shliaha PV, Miele M, Hendrickson RC, Jiang XJ, Helin K. H3k4me3 regulates rna polymerase ii promoter-proximal pause-release. Nature, 2023, 615(7951): 339-348.
pmid: 36859550 |
| [32] |
Zhao Y, Hu JT, Wu JJ, Li ZH. Chip-seq profiling of H3K4me3 and H3K27me3 in an invasive insect, bactrocera dorsalis. Front Genet, 2023, 14: 1108104.
pmid: 36911387 |
| [33] |
Ing-Simmons E, Seitan VC, Faure AJ, Flicek P, Carroll T, Dekker J, Fisher AG, Lenhard B, Merkenschlager M. Spatial enhancer clustering and regulation of enhancer- proximal genes by cohesin. Genome Res, 2015, 25(4): 504-513.
pmid: 25677180 |
| [34] |
Pradeepa MM. Causal role of histone acetylations in enhancer function. Transcription, 2017, 8(1): 40-47.
pmid: 27792455 |
| [35] |
Soldi M, Mari T, Nicosia L, Musiani D, Sigismondo G, Cuomo A, Pavesi G, Bonaldi T. Chromatin proteomics reveals novel combinatorial histone modification signatures that mark distinct subpopulations of macrophage enhancers. Nucleic Acids Res, 2017, 45(21): 12195-12213.
pmid: 28981749 |
| [36] |
Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell, 2013, 153(2): 307-319.
pmid: 23582322 |
| [37] |
Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA. Coactivator condensation at super-enhancers links phase separation and gene control. Science, 2018, 361(6400): eaar3958.
pmid: 29930091 |
| [38] |
Li MS, Huang HX, Wang BF, Jiang SS, Guo HZ, Zhu LQ, Wu SQ, Liu JF, Wang L, Lan XH, Zhang W, Zhu J, Li FX, Tan JY, Mao Z, Liu CQ, Ji JP, Ding JJ, Zhang K, Yuan J, Liu YZ, Ouyang H. Comprehensive 3d epigenomic maps define limbal stem/progenitor cell function and identity. Nat Commun, 2022, 13(1): 1293.
pmid: 35277509 |
| [39] |
Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet, 1979, 13: 319-353.
pmid: 94251 |
| [40] |
Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem, 1986, 55: 339-372.
pmid: 3527045 |
| [41] |
Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature, 1981, 289(5800): 751-758.
pmid: 7007895 |
| [42] |
Dorantes-Palma D, Pérez-Mora S, Azuara-Liceaga E, Pérez-Rueda E, Pérez-Ishiwara DG, Coca-González M, Medel-Flores MO, Gómez-García C. Screening and structural characterization of heat shock response elements (HSEs) in entamoeba histolytica promoters. Int J Mol Sci, 2024, 25(2): 1319.
pmid: 38279319 |
| [43] |
Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res, 2001, 29(14): 2905-2919.
pmid: 11452016 |
| [44] |
Pang BX, van Weerd JH, Hamoen FL, Snyder MP. Identification of non-coding silencer elements and their regulation of gene expression. Nat Rev Mol Cell Biol, 2023, 24(6): 383-395.
pmid: 36344659 |
| [45] |
Zabidi MA, Stark A. Regulatory enhancer-core-promoter communication via transcription factors and cofactors. Trends Genet, 2016, 32(12): 801-814.
pmid: 27816209 |
| [46] |
Gupte R, Nandu T, Kraus WL. Nuclear ADP-ribosylation drives IFNγ-dependent STAT1α enhancer formation in macrophages. Nat Commun, 2021, 12(1): 3931.
pmid: 34168143 |
| [47] |
Barshad G, Lewis JJ, Chivu AG, Abuhashem A, Krietenstein N, Rice EJ, Ma YT, Wang Z, Rando OJ, Hadjantonakis AK, Danko CG. RNA polymerase ii dynamics shape enhancer-promoter interactions. Nat Genet, 2023, 55(8): 1370-1380.
pmid: 37430091 |
| [48] |
Vihervaara A, Mahat DB, Guertin MJ, Chu TY, Danko CG, Lis JT, Sistonen L. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun, 2017, 8(1): 255.
pmid: 28811569 |
| [49] |
Theobald J, DiMario JX. Lineage-based primary muscle fiber type diversification independent of MEF2 and NFAT in chick embryos. J Muscle Res Cell Motil, 2011, 31(5-6): 369-381.
pmid: 21290171 |
| [50] |
Tai PW, Fisher-Aylor KI, Himeda CL, Smith CL, Mackenzie AP, Helterline DL, Angello JC, Welikson RE, Wold BJ, Hauschka SD. Differentiation and fiber type-specific activity of a muscle creatine kinase intronic enhancer. Skelet Muscle, 2011, 1: 25.
pmid: 21797989 |
| [51] |
Potthoff MJ, Wu H, Arnold MA, Shelton JM, Backs J, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest, 2007, 117(9): 2459-2467.
pmid: 17786239 |
| [52] |
Asfour H, Hirsinger E, Rouco R, Zarrouki F, Hayashi S, Swist S, Braun T, Patel K, Relaix F, Andrey G, Stricker S, Duprez D, Stantzou A, Amthor H. Inhibitory SMAD6 interferes with BMP-dependent generation of muscle progenitor cells and perturbs proximodistal pattern of murine limb muscles. Development, 2023, 150(11): dev201504.
pmid: 37272529 |
| [53] |
Bengtsen M, Winje IM, Eftestøl E, Landskron J, Sun CY, Nygård K, Domanska D, Millay DP, Meza-Zepeda LA, Gundersen K. Comparing the epigenetic landscape in myonuclei purified with a PCM1 antibody from a fast/glycolytic and a slow/oxidative muscle. PLoS Genet, 2021, 17(11): e1009907.
pmid: 34752468 |
| [54] |
Long KR, Su D, Li XK, Li HK, Zeng S, Zhang Y, Zhong ZN, Lin Y, Li XM, Lu L, Jin L, Ma JD, Tang QZ, Li MZ. Identification of enhancers responsible for the coordinated expression of myosin heavy chain isoforms in skeletal muscle. BMC Genomics, 2022, 23(1): 519.
pmid: 35842589 |
| [55] |
Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol, 1998, 14: 167-196.
pmid: 9891782 |
| [56] |
Liu L, Ding CY, Fu TT, Feng ZH, Lee JE, Xiao LW, Xu ZS, Yin YJ, Guo QQ, Sun ZC, Sun WP, Mao Y, Yang LK, Zhou Z, Zhou DX, Xu LL, Zhu ZZ, Qiu Y, Ge K, Gan ZJ. Histone methyltransferase MLL4 controls myofiber identity and muscle performance through MEF2 interaction. J Clin Invest, 2020, 130(9): 4710-4725.
pmid: 32544095 |
| [57] |
Liu YW, Fu Y, Yang YL, Yi GQ, Lian JM, Xie BK, Yao YL, Chen MY, Niu YC, Liu L, Wang LY, Zhang YS, Fan XH, Tang YJ, Yuan PX, Zhu M, Li QW, Zhang S, Chen Y, Wang BH, He JY, Lu D, Liachko I, Sullivan ST, Pang B, Chen YQ, He X, Li K, Tang ZL. Integration of multi-omics data reveals cis-regulatory variants that are associated with phenotypic differentiation of eastern from western pigs. Genet Sel Evol, 2022, 54(1): 62.
pmid: 36104777 |
| [58] |
O'Mahoney JV, Guven KL, Lin J, Joya JE, Robinson CS, Wade RP, Hardeman EC. Identification of a novel slow-muscle-fiber enhancer binding protein, MusTRD1. Mol Cell Biol, 1998, 18(11): 6641-6652.
pmid: 9774679 |
| [59] |
Corin SJ, Levitt LK, O'Mahoney JV, Joya JE, Hardeman EC, Wade R. Delineation of a slow-twitch-myofiber- specific transcriptional element by using in vivo somatic gene transfer. Proc Natl Acad Sci USA, 1995, 92(13): 6185-6189.
pmid: 7597099 |
| [60] |
Aharoni-Simon M, Hann-Obercyger M, Pen S, Madar Z, Tirosh O. Fatty liver is associated with impaired activity of PPARγ-coactivator 1α (PGC1α) and mitochondrial biogenesis in mice. Lab Invest, 2011, 91(7): 1018-1028.
pmid: 21464822 |
| [61] |
Heesch MW, Shute RJ, Kreiling JL, Slivka DR. Transcriptional control, but not subcellular location, of PGC-1α is altered following exercise in a hot environment. J Appl Physiol (1985), 2016, 121(3): 741-749.
pmid: 27445305 |
| [62] |
Lee YG, Song MY, Cho H, Jin JS, Park BH, Bae EJ. Limonium tetragonum promotes running endurance in mice through mitochondrial biogenesis and oxidative fiber formation. Nutrients, 2022, 14(19): 3904.
pmid: 36235564 |
| [63] |
Ting AKL, Siow NL, Kong LW, Tsim KWK. Transcriptional regulation of acetylcholinesterase-associated collagen colq in fast- and slow-twitch muscle fibers. Chem Biol Interact, 2005, 157-158: 63-70.
pmid: 16256971 |
| [64] |
Yuan RQ, Zhang JM, Wang YJ, Zhu XX, Hu SL, Zeng JH, Liang F, Tang QZ, Chen YS, Chen LX, Zhu W, Li MZ, Mo DL. Reorganization of chromatin architecture during prenatal development of porcine skeletal muscle. DNA Res, 2021, 28(2): dsab003.
pmid: 34009337 |
| [65] |
Li J, Lin Y, Li DY, He MN, Kui H, Bai JY, Chen ZY, Gou YW, Zhang JM, Wang T, Tang QZ, Kong FL, Jin L, Li MZ. Building haplotype-resolved 3d genome maps of chicken skeletal muscle. Adv Sci (Weinh), 2024, 11(24): e2305706.
pmid: 38582509 |
| [66] |
Dos Santos M, Shah AM, Zhang YC, Bezprozvannaya S, Chen KN, Xu L, Lin WC, McAnally JR, Bassel-Duby R, Liu N, Olson EN. Opposing gene regulatory programs governing myofiber development and maturation revealed at single nucleus resolution. Nat Commun, 2023, 14(1): 4333.
pmid: 37468485 |
| [67] | Zhang ZH, Lin SD, Huang X, Zhang XQ. Study on the regulations of Sox6 gene on skeletal muscle differentiation and muscle fiber types in chicken. China Poultry, 2024, 41(9): 8-14. |
| 张梓豪, 林树带, 黄幸, 张细权. Sox6基因调控鸡骨骼肌分化和肌纤维类型的研究. 中国家禽, 2019, 41(9): 8-14. | |
| [68] |
Yu MB, Feng YQ, Yan JM, Zhang XY, Tian Z, Wang T, Wang JJ, Shen W. Transcriptomic regulatory analysis of skeletal muscle development in landrace pigs. Gene, 2024, 915: 148407.
pmid: 38531491 |
| [69] |
Sadaki S, Fujita R, Hayashi T, Nakamura A, Okamura Y, Fuseya S, Hamada M, Warabi E, Kuno A, Ishii A, Muratani M, Okada R, Shiba D, Kudo T, Takeda S, Takahashi S. Large maf transcription factor family is a major regulator of fast type iib myofiber determination. Cell Rep, 2023, 42(4): 112289.
pmid: 36952339 |
| [70] |
Allen DL, Sartorius CA, Sycuro LK, Leinwand LA. Different pathways regulate expression of the skeletal myosin heavy chain genes. J Biol Chem, 2001, 276(47): 43524-43533.
pmid: 11551968 |
| [71] |
Maire P. New insights into adult muscle fiber-type diversity: involvement of six homeoproteins. Bull Acad Natl Med, 2015, 199(1): 21-31.
pmid: 27236875 |
| [72] |
Girgis J, Yang DB, Chakroun I, Liu YB, Blais A. Six1 promotes skeletal muscle thyroid hormone response through regulation of the mct10 transporter. Skelet Muscle, 2021, 11(1): 26.
pmid: 34809717 |
| [73] |
Concordet JP, Maire P, Kahn A, Daegelen D. A ubiquitous enhancer shared by two promoters in the human aldolase a gene. Nucleic Acids Res, 1991, 19(15): 4173-4180.
pmid: 1651479 |
| [74] |
Choi RCY, Ting AKL, Lau FTC, Xie HQ, Leung KW, Chen VP, Siow NL, Tsim KWK. Calcitonin gene-related peptide induces the expression of acetylcholinesterase- associated collagen colq in muscle: a distinction in driving two different promoters between fast- and slow-twitch muscle fibers. J Neurochem, 2007, 102(4): 1316-1328.
pmid: 17488278 |
| [75] | Lefaucheur L. Myofiber typing and pig meat production. Slovenski Veterinarski Zbornik, 2001, 38(1): 5-28. |
| [76] |
Gondret F, Combes S, Lefaucheur L, Lebret B. Effects of exercise during growth and alternative rearing systems on muscle fibers and collagen properties. Reprod Nutr Dev, 2005, 45(1): 69-86.
pmid: 15865057 |
| [77] |
Li CC, Wang YN, Sun XH, Yang JJ, Ren YC, Jia JR, Yang GS, Liao MZ, Jin JJ, Shi XE. Identification of different myofiber types in pigs muscles and construction of regulatory networks. BMC Genomics, 2024, 25(1): 400.
pmid: 38658807 |
| [78] |
Li S, Chen J, Wei PT, Zou TD, You JM. Fibroblast growth factor 21: a fascinating perspective on the regulation of muscle metabolism. Int J Mol Sci, 2023, 24(23): 16951.
pmid: 38069273 |
| [79] |
Liu XY, Wang YL, Hou LM, Xiong YZ, Zhao SH. Fibroblast growth factor 21 (FGF21) promotes formation of aerobic myofibers via the FGF21-SIRT1-AMPK-PGC1α pathway. J Cell Physiol, 2017, 232(7): 1893-1906.
pmid: 27966786 |
| [80] |
Lu TT, Zhu YF, Guo JL, Mo ZY, Zhou Q, Hu CY, Wang C. MDFI regulates fast-to-slow muscle fiber type transformation via the calcium signaling pathway. Biochem Biophys Res Commun, 2023, 671: 215-224.
pmid: 37307704 |
| [81] |
Dong C, Zhang XY, Liu KQ, Li BJ, Chao Z, Jiang AW, Li RY, Li PH, Liu HL, Wu WJ. Comprehensive analysis of porcine PROX1 gene and its relationship with meat quality traits. Animals (Basel), 2019, 9(10): 744.
pmid: 31569476 |
| [82] |
Kivelä R, Salmela I, Nguyen YH, Petrova TV, Koistinen HA, Wiener Z, Alitalo K. The transcription factor Prox1 is essential for satellite cell differentiation and muscle fibre-type regulation. Nat Commun, 2016, 7: 13124.
pmid: 27731315 |
| [83] |
Petchey LK, Risebro CA, Vieira JM, Roberts T, Bryson JB, Greensmith L, Lythgoe MF, Riley PR. Loss of Prox1 in striated muscle causes slow to fast skeletal muscle fiber conversion and dilated cardiomyopathy. Proc Natl Acad Sci USA, 2014, 111(26): 9515-9520.
pmid: 24938781 |
| [84] |
Guerfali I, Manissolle C, Durieux AC, Bonnefoy R, Bartegi A, Freyssenet D. Calcineurin A and CaMKIV transactivate PGC-1alpha promoter, but differentially regulate cytochrome c promoter in rat skeletal muscle. Pflugers Arch, 2007, 454(2): 297-305.
pmid: 17273866 |
| [85] |
Lin JD, Wu H, Tarr PT, Zhang CY, Wu ZD, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow- twitch muscle fibres. Nature, 2002, 418(6899): 797-801.
pmid: 12181572 |
| [86] |
Kim MS, Fielitz J, McAnally J, Shelton JM, Lemon DD, McKinsey TA, Richardson JA, Bassel-Duby R, Olson EN. Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Mol Cell Biol, 2008, 28(11): 3600-3609.
pmid: 18378694 |
| [1] | Meng Yuan, Hui Li, Shouzhi Wang. Massively parallel reporter assay: a novel technique for analyzing the regulation of gene expression [J]. Hereditas(Beijing), 2023, 45(10): 859-873. |
| [2] | Haidong Xu, Bolin Ning, Fang Mu, Hui Li, Ning Wang. Advances of functional consequences and regulation mechanisms of alternative cleavage and polyadenylation [J]. Hereditas(Beijing), 2021, 43(1): 4-15. |
| [3] | Jian Shi,Yanming Li,Xiangdong Fang. The mechanism and clinical significance of long noncoding RNA-mediated gene expression via nuclear architecture [J]. Hereditas(Beijing), 2017, 39(3): 189-199. |
| [4] | Chang Lu, Yinhua Huang. Progress in long non-coding RNAs in animals [J]. Hereditas(Beijing), 2017, 39(11): 1054-1065. |
| [5] | Fei Zhou, Shizhan Lu, Liang Gao, Juanjuan Zhang, Yongjun Lin. Plastid genome engineering: novel optimization strategies and applications [J]. HEREDITAS(Beijing), 2015, 37(8): 777-792. |
| [6] | Qiang Pu,Jia Luo,Linyuan Shen,Xuewei Li,Shunhua Zhang,Li Zhu. Application of modification-specific proteomics in the meat-quality study [J]. HEREDITAS(Beijing), 2015, 37(4): 327-335. |
| [7] | Xiaoqing Huang,Dandan Li,Juan Wu. Long non-coding RNAs in plants [J]. HEREDITAS(Beijing), 2015, 37(4): 344-359. |
| [8] | SHEN Lin-Yuan ZHANG Shun-Hua WU Ze-Hui ZHENG Meng-Yue LI Xue-Wei, ZHU Li. The influence of satellite cells on meat quality and its differential regulation [J]. HEREDITAS, 2013, 35(9): 1081-1086. |
| [9] | XIA Tian, XIAO Bing-Xiu, GUO Jun-Ming. Acting mechanisms and research methods of long noncoding RNAs [J]. HEREDITAS, 2013, 35(3): 269-280. |
| [10] | QIN Dan, XU Cun-Shuan. Characterization and identification of functional elements in non-coding DNA sequences [J]. HEREDITAS, 2013, 35(11): 1253-1264. |
| [11] | YANG Fan, WANG Qiong-Ping, HE Kan, WANG Ming-Hui, PAN Yu-Chun. Association between gene polymorphisms of propanoate metabolism pathway and meat quality as well as carcass traits in pigs [J]. HEREDITAS, 2012, 34(7): 872-878. |
| [12] | LUO Mao, ZHANG Zhi-Ming, GAO Jian, ZENG Xing, PAN Guang-Tang. The role of miR319 in plant development regulation [J]. HEREDITAS, 2011, 33(11): 1203-1211. |
| [13] | . Molecular mechanisms of regulation of estrogen receptor a expression level in breast cancer [J]. HEREDITAS, 2010, 32(3): 191-197. |
| [14] | DING Yan-Fei, WANG Guang-Yue, FU E-Ping, SHU Cheng. The role of miR398 in plant stress responses [J]. HEREDITAS, 2010, 32(2): 129-134. |
| [15] | SU Hui-Zhao, XIANG Zhi-Jiao, BANG Fang-Yi, LI Rui-Fang, AN Shi-Qi, LIU Guang-Chao, TANG Ji-Liang. Identification of a gene involved in the expression of the pathogenicity-related gene XC3814 in Xanthomonas campestris [J]. HEREDITAS, 2010, 32(1): 81-86. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||