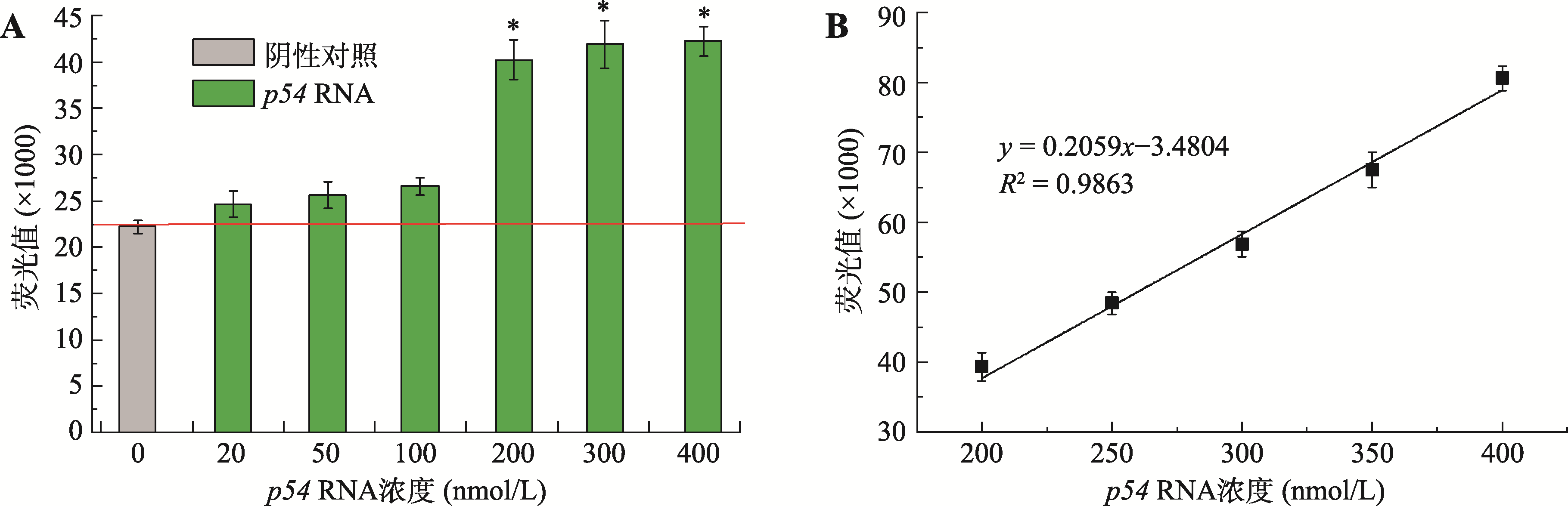
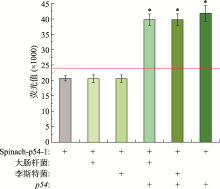
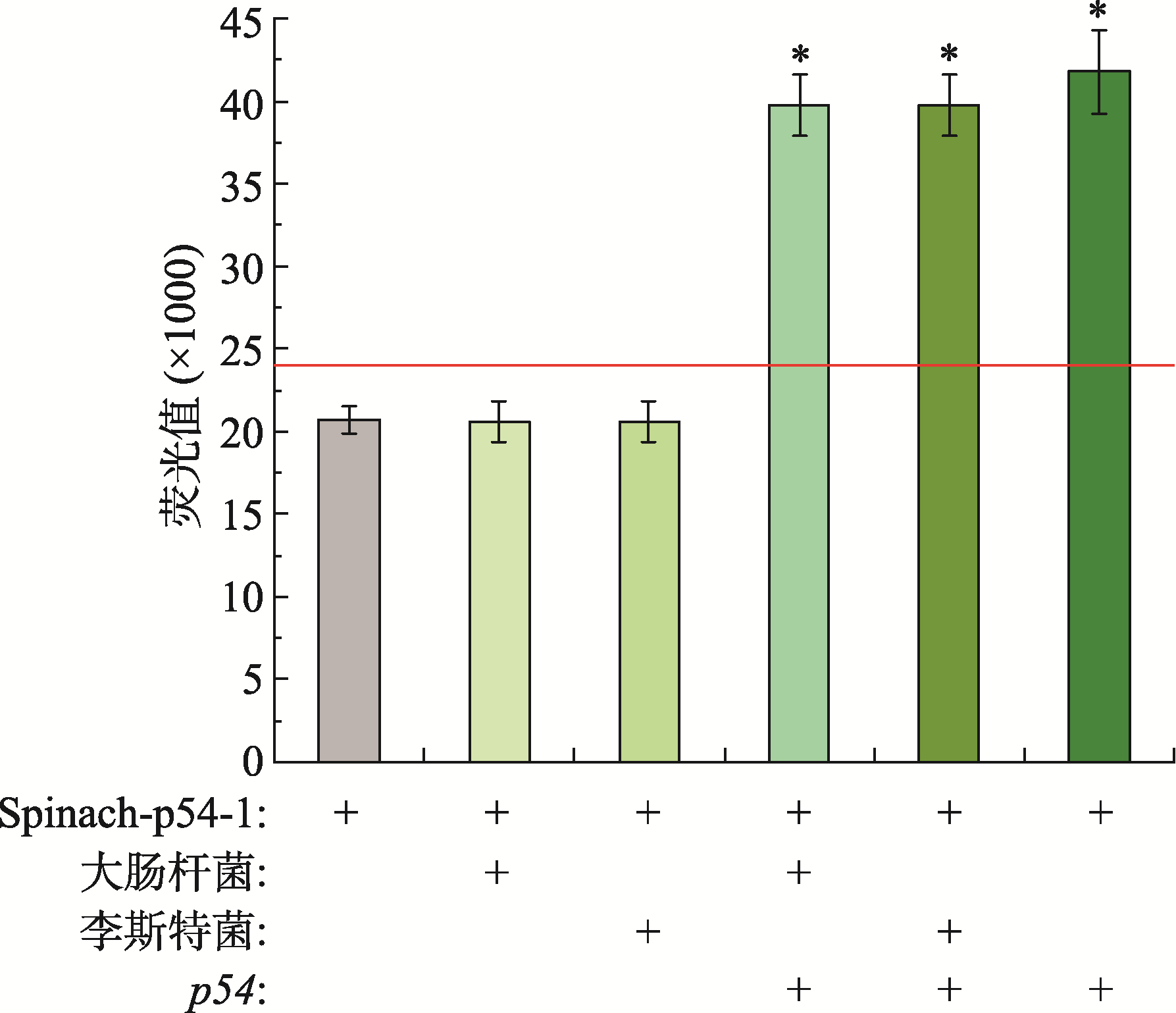
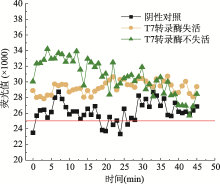
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (12): 1170-1178.doi: 10.16288/j.yczz.21-335
• Technique and Method • Previous Articles Next Articles
Design and application of lightening Spinach-p54 RNA apatmer to detect African swine fever virus
Chengcheng Han1,2( ), Kai Xia1,3, Ruying Gong1, Xuhan Wu1, Lei Zhang1,2(
), Kai Xia1,3, Ruying Gong1, Xuhan Wu1, Lei Zhang1,2( ), Xinle Liang1,2(
), Xinle Liang1,2( )
)
- 1. School of Food Science and Biotechnology, Zhejiang Gongshang University, Hangzhou 310018, China
2. Institute of Food Biotechnology, Zhejiang Gongshang University, Hangzhou 310018, China
3. School of Biological and Chemical Engineering, Zhejiang University of Science and Technology, Hangzhou 310023, China
-
Received:2021-09-21Revised:2021-11-30Online:2021-12-20Published:2021-12-10 -
Contact:Zhang Lei,Liang Xinle E-mail:707884766@qq.com;zhanglei@zjsu.edu.cn;dbiot@mail.zjgsu.edu.cn
Cite this article
Chengcheng Han, Kai Xia, Ruying Gong, Xuhan Wu, Lei Zhang, Xinle Liang. Design and application of lightening Spinach-p54 RNA apatmer to detect African swine fever virus[J]. Hereditas(Beijing), 2021, 43(12): 1170-1178.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
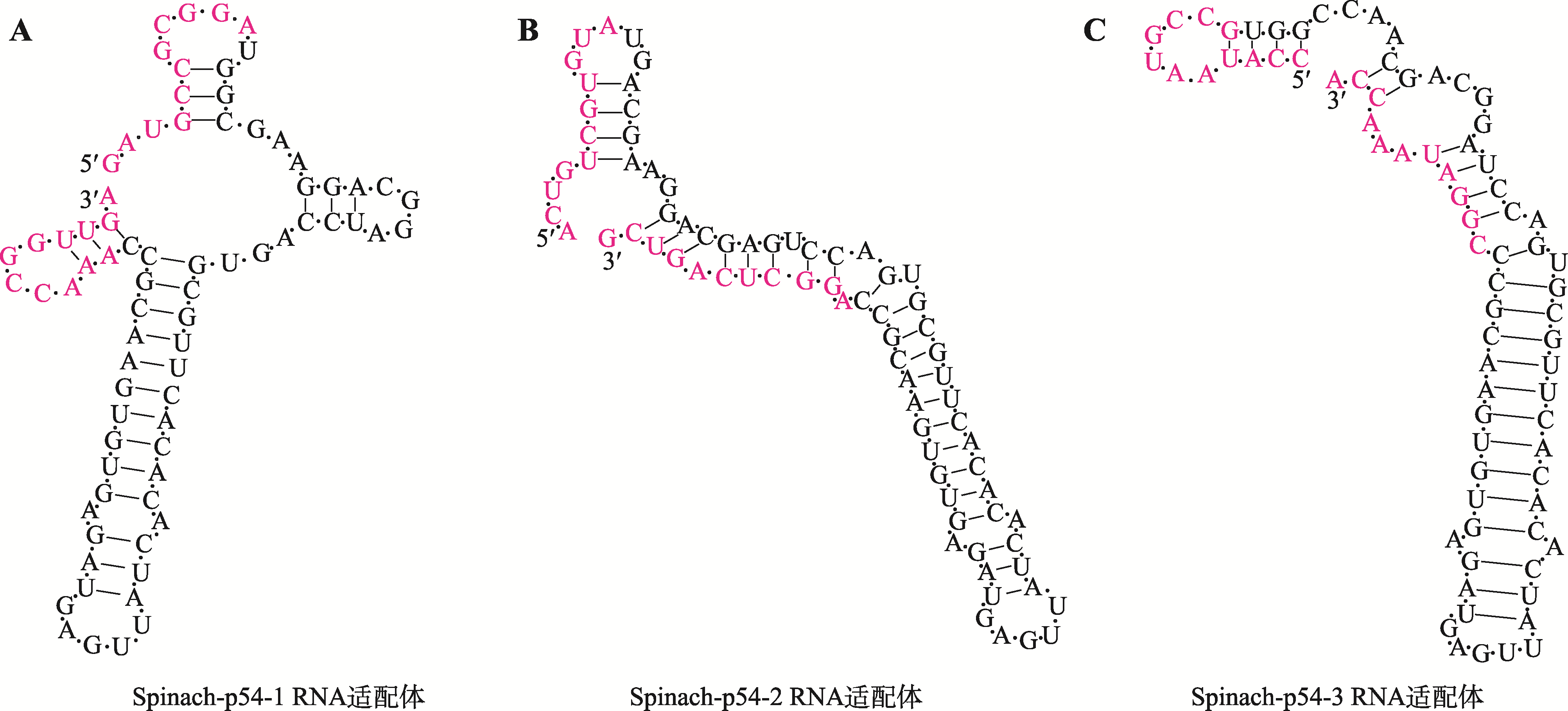
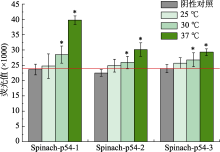
Table 1
Oligonucleotides used in this study"
| 适配体名称 | 序列(5′→3′) |
|---|---|
| Spinach-p54-1 | GATGCCGCGGATGGCGAAGGACGGGTCCAGTGCGTTCGCGCACTGTTGAGTAGAGTGTGAGCGCCAAACCGGTTGA |
| Spinach-p54-2 | ACTGTCGTGTATGGCGAAGGACGGGTCCAGTGCGTTCGCGCACTGTTGAGTAGAGTGTGAGCGCCAGGCTCAGTCG |
| Spinach-p54-3 | CCATAATGCCGTGGCGAAGGACGGGTCCAGTGCGTTCGCGCACTGTTGAGTAGAGTGTGAGCGCCCGGATAAACCG |
| [1] | Normile D. Arrival of deadly pig disease could spell disaster for China. Science, 2018,361(6404):741. |
| [2] | Ma J, Chen H, Gao X, Xiao JH, Wang HB. African swine fever emerging in China: distribution characteristics and high-risk areas. Prev Vet Med, 2020,175:104861. |
| [3] | Galindo I, Alonso C. African swine fever virus: a review. Viruses, 2017,9(5):103. |
| [4] | Giudici SD, Franzoni G, Bonelli P, Bacciu D, Sanna G, Angioi PP, Ledda M, Pilo G, Nicolussi P, Oggiano A. Interaction of historical and modern Sardinian African swine fever viruses with porcine and wild-boar monocytes and monocyte-derived macrophages. Arch Virol, 2019,164(3):739-745. |
| [5] | Yang F, Cheng LQ, Xu XG, Zhang WM, Zhang Q. Establishment and preliminary application of a real-time fluorescence quantitative PCR for CSFV detection. Prog Vet Med, 2021,42(1):1-5. |
| 杨峰, 陈立强, 许信刚, 张为民, 张琪. 猪瘟病毒实时荧光定量PCR检测方法的建立及初步应用. 动物医学进展, 2021,42(1):1-5. | |
| [6] | Zhu YS, Shao N, Chen JW, Qi WB, Li Y, Liu P, Chen YJ, Bian SY, Zhang Y, Tao SC. Multiplex and visual detection of African swine fever virus (ASFV) based on Hive-Chip and direct loop-mediated isothermal amplification. Anal Chim Acta, 2020,1140:30-40. |
| [7] | Guinat C, Gogin A, Blome S, Kei G, Pollin R, Pfeiffer DU, Dixon L. Transmission routes of African swine fever virus to domestic pigs: current knowledge and future research directions. Vet Rec, 2016,178(11):262-267. |
| [8] | Wang ZY, Dan K, Shi YG, Kan SF, Zhong LX, He QH. Research progress on rapid detection methods of African swine fever virus. Food Safe Qual Detec Technol, 2020,11(10):3215-3220. |
| 王梓莹, 但琨, 施远国, 阚式绂, 钟雷响, 何庆华. 非洲猪瘟病毒的快速检测方法研究进展. 食品安全质量检测学报, 2020,11(10):3215-3220. | |
| [9] | Cui BB, Li T, Qiu SY, Mei L, Han XQ, Wu SQ. Establishment of a real-time PCR for detection of African swine fever virus E184L gene. Prog Vet Med, 2020,41(1):8-14. |
| 崔贝贝, 李霆, 仇松寅, 梅琳, 韩雪清, 吴绍强, 林祥梅. 基于E184L基因的非洲猪瘟病毒实时荧光定量PCR检测方法的建立. 动物医学进展, 2020,41(1):8-14. | |
| [10] | Wang SJ, Liu MF, Yan RQ, Ban FG, Zhao XL, Ma ZY, Wang HJ, Wang C, Zhao MJ. Establishment and application of real-time quantitative PCR assay for detection of classical Swine fever virus. Chin Anim Husb Vet Med, 2017,44(7):2112-2118. |
| 王淑娟, 刘梅芬, 闫若潜, 班付国, 赵雪丽, 马震原, 王华俊, 王翠, 赵明军. 猪瘟病毒实时荧光定量PCR检测方法的建立与应用. 中国畜牧兽医, 2017,44(7):2112-2118. | |
| [11] | Yan L, Song S, Zhang JH, Tang XL, Zhang HY, Guo KP. Establishment and application of droplet digital PCR for detection of African swine fever virus. Acta Laser Biol Sin, 2020,29(4):344-351. |
| 严礼, 宋晟, 张继红, 唐小兰, 张海韵, 郭焜鹏. 非洲猪瘟病毒微滴式数字PCR检测方法的建立与应用. 激光生物学报, 2020,29(4):344-351. | |
| [12] | Bergeron HC, Glas PS, Schumann KR. Diagnostic specificity of the African swine fever virus antibody detection enzyme-linked immunosorbent assay in feral and domestic pigs in the United States. Transbound Emerg Dis, 2017,64(6):1665-1668. |
| [13] | Huang J, Huang JJ, Cheng XF, Li GX, Gao F, Tong GZ. Expression of p30 protein of African swine fever virus in prokaryotic cells and preparation of polyclonal antibodies. Chin J Vet Parasitol, 2020,28(5):37-41. |
| 黄剑, 徐晶晶, 程雪飞, 李国新, 高飞, 童光志. 非洲猪瘟病毒p30蛋白的原核表达及其多克隆抗体制备. 中国动物传染病学报, 2020,28(5):37-41. | |
| [14] | Liu XT, Wang ZY, Xin T, Guo XY, Jiang YT, Cui S, Yu HN, Zhu HF, Jia H. Prokaryotic expression and polyclonal antibody preparation and identification of African swine fever virus B438L protein. Chin Anim Husb Vet Med, 2021,48(3):991-1000. |
| 刘雪婷, 王召阳, 鑫婷, 郭晓宇, 姜一曈, 崔帅, 于海男, 朱鸿飞, 贾红. 非洲猪瘟病毒B438L蛋白的原核表达及其多克隆抗体的制备与鉴定. 中国畜牧兽医, 2021,48(3):991-1000. | |
| [15] | Bouhedda F, Autour A, Ryckelynck M. Light-up RNA aptamers and their cognate fluorogens: from their development to their applications. Int J Mol Sci, 2017,19(1):44. |
| [16] | Rogers TA, Andrews GE, Jaeger L, Grabow WW. Fluorescent monitoring of RNA assembly and processing using the split-spinach aptamer. ACS Synth Biol, 2015,4(2):162-166. |
| [17] | Chandler M, Lyalina T, Halman J, Rackley L, Lee L, Dang D, Ke WN, Sajja S, Woods S, Acharya S, Baumgarten E, Christopher J, Elshalia E, Hrebien G, Kublank K, Saleh S, Stallings B, Tafere M, Striplin C, Afonin KA. Broccoli fluorets: split aptamers as a user-friendly fluorescent toolkit for dynamic RNA nanotechnology. Molecules, 2018,23(12):3178. |
| [18] | Kikuchi N, Kolpashchikov DM. Split Spinach aptamer for highly selective recognition of DNA and RNA at ambient temperatures. ChemBioChem, 2016,17(17):1589-1592. |
| [19] | Ausländer S, Fuchs D, Hürlemann S, Ausländer D, Fussenegger M. Engineering a ribozyme cleavage-induced split fluorescent aptamer complementation assay. Nucleic Acids Res, 2016,44(10):e94. |
| [20] | Ong WQ, Citron YR, Sekine S, Huang B. Live cell imaging of endogenous mRNA using RNA-based fluorescence “Turn-On” probe. ACS Chem Biol, 2017,12(1):200-205. |
| [21] | Bhadra S, Ellington AD. Design and application of cotranscriptional non-enzymatic RNA circuits and signal transducers. Nucleic Acids Res, 2014,42(7):e58. |
| [22] | Gui HL, Jin QR, Zhang YJ, Wang XD, Yang YC, Shao CY, Cheng CY, Wei FF, Yang Y, Yang MH, Song HH. Development of an aptamer/fluorescence dye PicoGreen-based method for detection of fumonisin B1. Chin J Biotech, 2015,31(9):1393-1400. |
| 桂海娈, 金庆日, 张亚军, 王晓杜, 杨永春, 邵春艳, 程昌勇, 卫芳芳, 杨杨, 杨梦华, 宋厚辉. 基于荧光染料PicoGreen和核酸适配体的伏马毒素B1检测方法. 生物工程学报, 2015,31(9):1393-1400. | |
| [23] | Fernandez-Millan P, Autour A, Ennifar E, Westhof E, Ryckelynck M. Crystal structure and fluorescence properties of the iSpinach aptamer in complex with DFHBI. RNA, 2017,23(12):1788-1795. |
| [24] | Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, Rice PA, Piccirilli JA. A G-quadruplex- containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol, 2014,10(8):686-691. |
| [25] | Truong L, Ferré-D'Amaré AR. From fluorescent proteins to fluorogenic RNAs: tools for imaging cellular macromolecules. Protein Sci, 2019,28(8):1374-1386. |
| [26] | Matsunaga KI, Kimoto M, Lim VW, Thein TL, Vasoo S, Leo YS, Sun W, Hirao I. Competitive ELISA for a serologic test to detect dengue serotype-specific anti-NS1 IgGs using high-affinity UB-DNA aptamers. Sci Rep, 2021,11(1):18000. |
| [27] | Chen H, Park SG, Choi N, Kwon HJ, Kang T, Lee MK, Choo J. Sensitive detection of SARS-CoV-2 using a SERS-based aptasensor. ACS Sens, 2021,6(6):2378-2385. |
| [28] | Jeng SCY, Chan HHY, Booy EP, McKenna SA, Unrau PJ. Fluorophore ligand binding and complex stabilization of the RNA Mango and RNA Spinach aptamers. RNA, 2016,22(12):1884-1892. |
| [29] | Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc, 2014,136(46):16299-16308. |
| [30] | Parra-Guardado AL, Sweeney CL, Hayes EK, Trueman BF, Huang Y, Jamieson RC, Rand JL, Gagnon GA, Stoddart AK. Development of a rapid pre-concentration protocol and a magnetic beads-based RNA extraction method for SARS-CoV-2 detection in raw municipal wastewater. Environ Sci: Water Res Technol, 2021. |
| [31] | Modh H, Scheper T, Walter JG. Aptamer-modified magnetic beads in biosensing. Sensors (Basel), 2018,18(4):1041. |
| [32] | Wei Y, Li K, Lu DR, Zhu HX. Optimization of CUT&Tag product recovery and library construction method. Hereditas(Beijing), 2021,43(4):362-374. |
| 韦晔, 李科, 卢大儒, 朱化星. CUT&Tag产物回收和建库方法的优化. 遗传, 2021,43(4):362-374. | |
| [33] | Wang Y, Xu LZ, Noll L, Stoy C, Porter E, Fu JP, Feng Y, Peddireddi L, Liu XM, Dodd KA, Jia W, Bai JF. Development of a real-time PCR assay for detection of African swine fever virus with an endogenous internal control. Transbound Emerg Dis, 2020,67(6):2446-2454. |
| [34] | Fu JY, Zhang YP, Cai G, Meng G, Shi SB. Rapid and sensitive RPA-Cas12a-fluorescence assay for point-of-care detection of African swine fever virus. PLoS One, 2021,16(7):e0254815. |
| [35] | Wang XJ, Ji PP, Fan HY, Dang L, Wan WW, Liu SY, Li YH, Yu WX, Li XY, Ma XD, Ma X, Zhao Q, Huang XX, Liao M. CRISPR/Cas12a technology combined with immunochromatographic strips for portable detection of African swine fever virus. Commun Biol, 2020,3(1):62. |
| [1] | Bingzheng Wang, Chao Zhang, Jiali Zhang, Jin Sun. Conditional editing of the Drosophila melanogaster genome using single transcripts expressing Cas9 and sgRNA [J]. Hereditas(Beijing), 2023, 45(7): 593-601. |
| [2] | Penghui Song, Lijuan Ma, Dong Yan. Exon junction complex modulates the formation of the m6A epitranscriptome [J]. Hereditas(Beijing), 2023, 45(6): 464-471. |
| [3] | Chengxian Wang, Yikang S. Rong, Min Cui. The molecular mechanism of Drosophila restricting telomeric transposons [J]. Hereditas(Beijing), 2023, 45(3): 221-228. |
| [4] | Xi Han, Fucheng Luo. Application of single-cell RNA sequencing in probing oligodendroglia heterogeneity and neurological disorders [J]. Hereditas(Beijing), 2023, 45(3): 198-211. |
| [5] | Biyuan Li, Yanting Zhao, Zhichen Yue, Juanli Lei, Qizan Hu, Peng Tao. Identification of DELLA gene family in head cabbage and analysis of mRNA transport in the heterograft [J]. Hereditas(Beijing), 2023, 45(2): 156-164. |
| [6] | Fengyu Sun, Qianghua Xu. Research progress of microRNAs involved in hematopoiesis [J]. Hereditas(Beijing), 2022, 44(9): 756-771. |
| [7] | Siyuan Xu, Jia Shou, Qiang Wu. Additional evidence of HS5-1 enhancer eRNA PEARL for protocadherin alpha gene regulation [J]. Hereditas(Beijing), 2022, 44(8): 695-764. |
| [8] | Xue Lv, Bangjie Li, Hanmei Xu. Research progress on high throughput discovery and functional verification of functional micropeptides [J]. Hereditas(Beijing), 2022, 44(6): 478-490. |
| [9] | Juan Wang, Yuening Yang, Weilan Piao, Hua Jin. Uridylation: a vital way for cellular RNA surveillance [J]. Hereditas(Beijing), 2022, 44(6): 449-465. |
| [10] | Wandi Xiong, Kaiyu Xu, Lin Lu, Jiali Li. Research progress on lncRNAs in Alzheimer’s disease [J]. Hereditas(Beijing), 2022, 44(3): 189-197. |
| [11] | Min Cheng, Jing Zhang, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of the hypoxia-associated long non-coding RNA signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2022, 44(2): 153-167. |
| [12] | Liping Tan, Rufei Gao, Xin Yin, Xuemei Chen, Fangfang Li, Liu Yuan, Junlin He. The expression of lincRNA AC027700.1 in mouse decidualization [J]. Hereditas(Beijing), 2022, 44(2): 168-177. |
| [13] | Yuting Han, Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni. The cutting edge of gene regulation approaches in model organism Drosophila [J]. Hereditas(Beijing), 2022, 44(1): 3-14. |
| [14] | Kenao Lv, Xuefeng Pan. Progress on the mechanistic research of the trinucleotide repeat instabilities underlying human neurodegenerative diseases [J]. Hereditas(Beijing), 2021, 43(9): 835-848. |
| [15] | Weilu Zhang, Qiying Leng, Jiahui Zheng, Ali Hassan Nawaz, Zhenhai Jiao, Fujian Wang, Li Zhang. Cloning and expression of circular transcript of mouse growth hormone receptor gene [J]. Hereditas(Beijing), 2021, 43(9): 890-900. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||