Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (1): 3-14.doi: 10.16288/j.yczz.21-347
• Orginal Articles • Previous Articles Next Articles
The cutting edge of gene regulation approaches in model organism Drosophila
Yuting Han( ), Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni(
), Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni( )
)
- Gene Regulatory Lab, School of Medicine, Tsinghua University, Beijing 100084, China
-
Received:2021-10-07Revised:2021-12-21Online:2022-01-20Published:2022-01-04 -
Contact:Ni Jianquan E-mail:hanyt19@mails.tsinghua.edu.cn;nijq@mail.tsinghua.edu.cn -
Supported by:Supported by the National Natural Science Foundation of China Nos(20181300988);Supported by the National Natural Science Foundation of China Nos(20201300797);the National Key Technology Research and Development Program of the Ministry of Science and Technology of the People’s Republic of China No(2016YFE0113700)
Cite this article
Yuting Han, Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni. The cutting edge of gene regulation approaches in model organism Drosophila[J]. Hereditas(Beijing), 2022, 44(1): 3-14.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Esquerda-Canals G, Montoliu-Gaya L, Güell-Bosch J, Villegas S. Mouse models of Alzheimer's disease. J Alzheimers Dis, 2017, 57(4):1171-1183.
doi: 10.3233/JAD-170045 pmid: 28304309 |
| [2] |
Jin HL, He R, Oyoshi M, Geha RS. Animal models of atopic dermatitis. J Invest Dermatol, 2009, 129(1):31-40.
doi: 10.1038/jid.2008.106 |
| [3] |
High KA, Roncarolo MG. Gene therapy. N Engl J Med, 2019, 381(5):455-464.
doi: 10.1056/NEJMra1706910 |
| [4] |
Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature, 1980, 287(5785):795-801.
doi: 10.1038/287795a0 |
| [5] |
Hoffmann JA. The immune response ofDrosophila. Nature, 2003, 426(6962):33-38.
doi: 10.1038/nature02021 |
| [6] |
Konopka RJ, Benzer S. Clock mutants ofDrosophila melanogaster. Proc Natl Acad Sci USA, 1971, 68(9):2112-2116.
doi: 10.1073/pnas.68.9.2112 |
| [7] |
Housden BE, Muhar M, Gemberling M, Gersbach CA, Stainier DYR, Seydoux G, Mohr SE, Zuber J, Perrimon N. Loss-of-function genetic tools for animal models: cross-species and cross-platform differences. Nat Rev Genet, 2017, 18(1):24-40.
doi: 10.1038/nrg.2016.118 |
| [8] |
Xu RG, Wang X, Shen D, Sun J, Qiao HH, Wang F, Liu LP, Ni JQ. Perspectives on gene expression regulation techniques inDrosophila. J Genet Genomics, 2019, 46(4):213-220.
doi: 10.1016/j.jgg.2019.03.006 |
| [9] |
Xu J, Ren XJ, Sun J, Wang X, Qiao HH, Xu B-W, Liu LP, Ni JQ. A toolkit of CRISPR-based genome editing systems inDrosophila. J Genet Genomics, 2015, 42(4):141-149.
doi: 10.1016/j.jgg.2015.02.007 |
| [10] | Mohr SE, Smith JA, Shamu CE, Neumüller RA, Perrimon N. RNAi screening comes of age: improved techniques and complementary approaches. Nat Rev Mol Cell Biol, 2014, 15(9):591-600. |
| [11] |
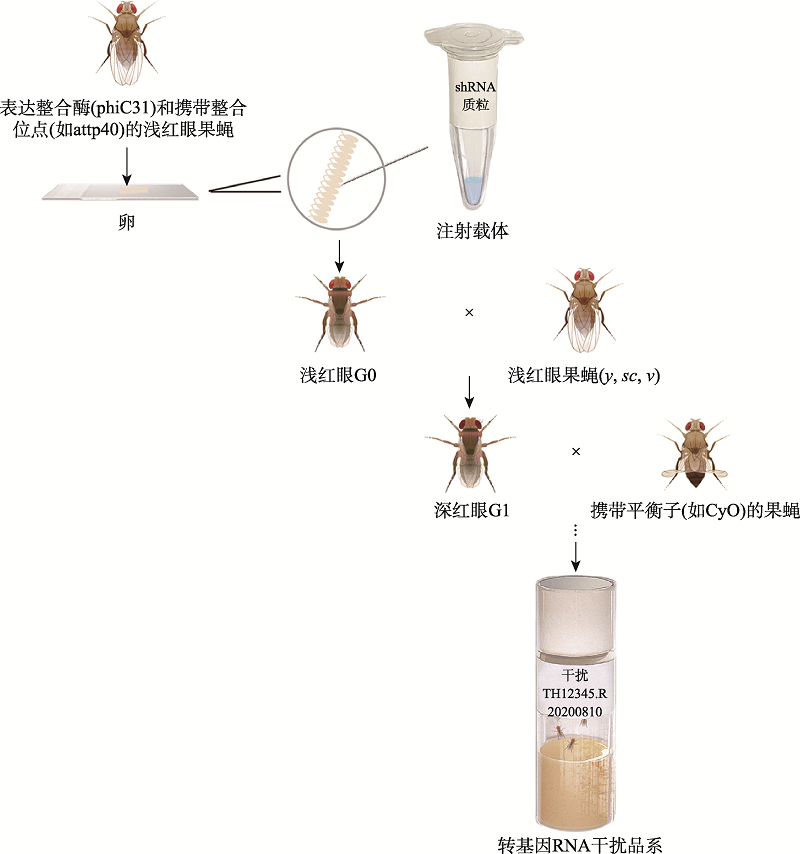
Qiao HH, Wang F, Xu RG, Sun J, Zhu R, Mao D, Ren XJ, Wang X, Jia Y, Peng P, Shen D, Liu LP, Chang ZJ, Wang GR, Li S, Ji JY, Liu QF, Ni JQ. An efficient and multiple target transgenic RNAi technique with low toxicity inDrosophila. Nat Commun, 2018, 9(1):4160.
doi: 10.1038/s41467-018-06537-y |
| [12] | Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods, 2011, 8(5):405-407. |
| [13] |
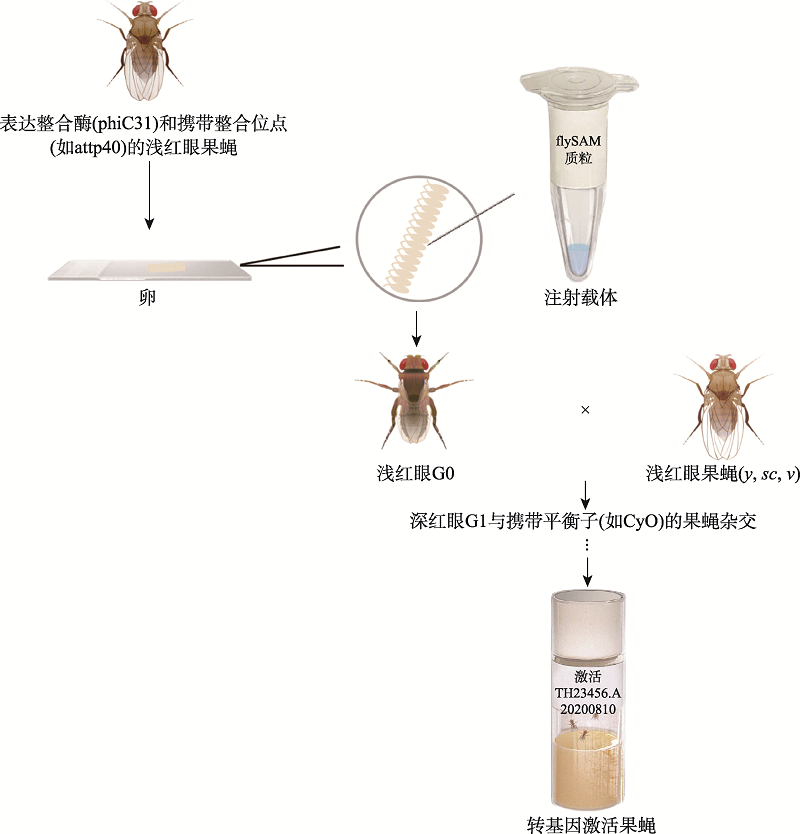
Jia Y, Xu RG, Ren XJ, Ewen-Campen B, Rajakumar R, Zirin J, Yang-Zhou D, Zhu RB, Wang F, Mao D, Peng P, Qiao HH, Wang X, Liu LP, Xu BW, Ji JY, Liu QF, Sun J, Perrimon N, Ni JQ. Next-generation CRISPR/Cas9 transcriptional activation in Drosophila using flySAM. Proc Natl Acad Sci USA, 2018, 115(18):4719-4724.
doi: 10.1073/pnas.1800677115 |
| [14] | Anderson P. Mutagenesis. Methods Cell Biol, 1995, 48:31-58. |
| [15] |
Ren XJ, Holsteens K, Li HY, Sun J, Zhang YF, Liu LP, Liu QF, Ni JQ. Genome editing in Drosophila melanogaster: from basic genome engineering to the multipurpose CRISPR-Cas9 system. Sci China Life Sci, 2017, 60(5):476-489.
doi: 10.1007/s11427-017-9029-9 |
| [16] |
Bak RO, Gomez-Ospina N, Porteus MH. Gene editing on center stage. Trends Genet, 2018, 34(8):600-611.
doi: 10.1016/j.tig.2018.05.004 |
| [17] |
Gaj T, Gersbach CA, Barbas III CF. ZFN, TALEN, CRISPR/Cas-based methods for genome engineering. Trends Biotechnol, 2013, 31(7):397-405.
doi: 10.1016/j.tibtech.2013.04.004 |
| [18] |
Musunuru K. The hope and hype of CRISPR-Cas9 genome editing: a review. JAMA Cardiol, 2017, 2(8):914-919.
doi: 10.1001/jamacardio.2017.1713 pmid: 28614576 |
| [19] |
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell, 2014, 157(6):1262-1278.
doi: 10.1016/j.cell.2014.05.010 |
| [20] |
Ren XJ, Sun J, Housden BE, Hu YH, Roesel C, Lin SL, Liu LP, Yang ZH, Mao D, Sun LZ, Wu QJ, Ji JY, Xi JZ, Mohr SE, Xu J, Perrimon N, Ni JQ. Optimized gene editing technology forDrosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci USA, 2013, 110(47):19012-19017.
doi: 10.1073/pnas.1318481110 |
| [21] | Peng P, Wang X, Shen D, Sun J, Jia Y, Xu RG, Zhu LF, Ni JQ. CRISPR-Cas9 mediated genome editing inDrosophila. Bio Protoc, 2019, 9(2):e3141. |
| [22] |
Ren XJ, Yang ZH, Xu J, Sun J, Mao DC, Hu YH, Yang SJ, Qiao HH, Wang X, Hu Q, Deng P, Liu LP, Ji JY, Li JB, Ni JQ. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters inDrosophila. Cell Rep, 2014, 9(3):1151-1162.
doi: 10.1016/j.celrep.2014.09.044 |
| [23] |
Port F, Chen H, Lee T, Bullock S. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering inDrosophila. Proc Natl Acad Sci USA, 2014, 111(29):E2967-E2976.
doi: 10.1073/pnas.1405500111 |
| [24] |
Xue ZY, Wu MH, Wen KJ, Ren MD, Long L, Zhang XD, Gao GJ. CRISPR/Cas9 mediates efficient conditional mutagenesis inDrosophila. G3 (Bethesda), 2014, 4(11):2167-73.
doi: 10.1534/g3.114.014159 |
| [25] |
Liu QH, Paroo Z. Biochemical principles of small RNA pathways. Annu Rev Biochem, 2010, 79:295-319.
doi: 10.1146/biochem.2010.79.issue-1 |
| [26] |
Williams L, Carles CC, Osmont KS, Fletcher JC. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets theArabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci USA, 2005, 102(27):9703-9708.
doi: 10.1073/pnas.0504029102 |
| [27] |
Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 1993, 118(2):401-415.
pmid: 8223268 |
| [28] |
McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction inDrosophila. Science, 2003, 302(5651):1765-1768.
pmid: 14657498 |
| [29] |
Lam G, Thummel CS. Inducible expression of double- stranded RNA directs specific genetic interference inDrosophila. Curr Biol, 2000, 10(16):957-963.
pmid: 10985382 |
| [30] |
Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol, 2000, 18(8):896-898.
pmid: 10932163 |
| [31] |
Ni JQ, Markstein M, Binari R, Pfeiffer B, Liu LP, Villalta C, Booker M, Perkins L, Perrimon N. Vector and parameters for targeted transgenic RNA interference inDrosophila melanogaster. Nat Methods, 2008, 5(1):49-51.
doi: 10.1038/nmeth1146 |
| [32] | Wang F, Qiao HH, Xu RG, Sun J, Zhu RB, Mao DC, Ni JQ. pNP transgenic RNAi system manual in Drosophila. Bio Protoc, 2019, 9(3):e3158. |
| [33] |
Pfeiffer BD, Jenett A, Hammonds AS, Ngo T-TB, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics inDrosophila. Proc Natl Acad Sci USA, 2008, 105(28):9715-9720.
doi: 10.1073/pnas.0803697105 |
| [34] |
Rørth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci USA, 1996, 93(22):12418-12422.
doi: 10.1073/pnas.93.22.12418 |
| [35] |
Wei P, Xue W, Zhao Y, Ning G, Wang JW. CRISPR-based modular assembly of a UAS-cDNA/ORF plasmid library for more than 5500Drosophila genes conserved in humans. Genome Res, 2020, 30(1):95-106.
doi: 10.1101/gr.250811.119 |
| [36] |
Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. TheStreptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res, 2011, 39(21):9275-9282.
doi: 10.1093/nar/gkr606 pmid: 21813460 |
| [37] |
Lin SL, Ewen-Campen B, Ni XC, Housden BE, Perrimon N. In vivo transcriptional activation using CRISPR/Cas9 in Drosophila. Genetics, 2015, 201(2):433-442.
doi: 10.1534/genetics.115.181065 |
| [38] |
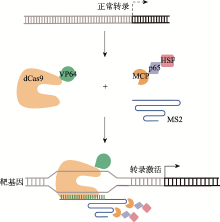
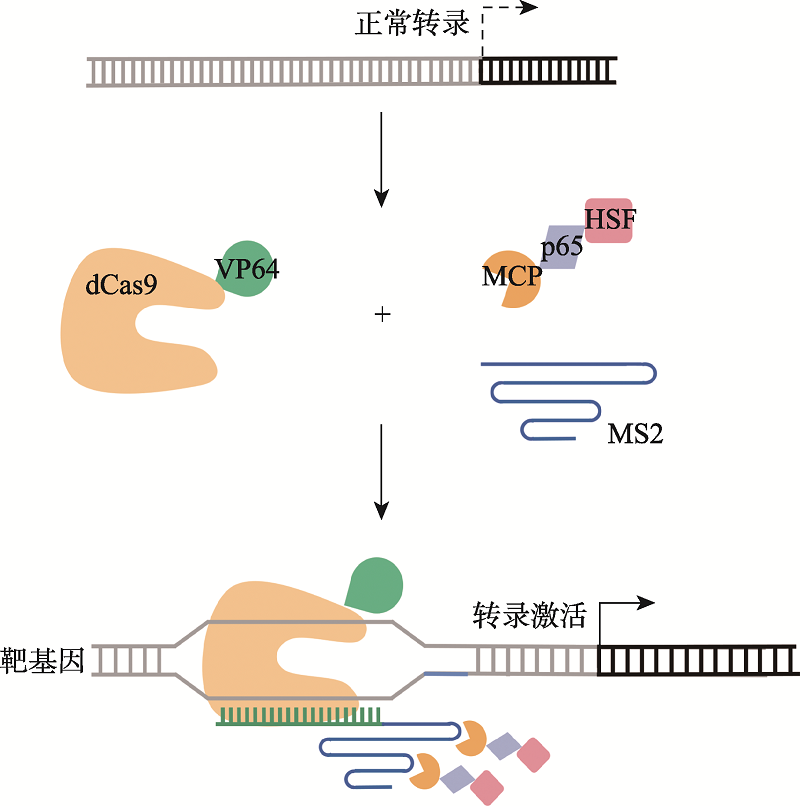
Ewen-Campen B, Yang-Zhou D, Fernandes VR, González DP, Liu LP, Tao R, Ren XJ, Sun J, Hu YH, Zirin J, Mohr SE, Ni JQ, Perrimon N. Optimized strategy for in vivo Cas9-activation in Drosophila. Proc Natl Acad Sci USA, 2017, 114(35):9409-9414.
doi: 10.1073/pnas.1707635114 |
| [39] |
Jia Y, Shen D, Wang X, Sun J, Peng P, Xu RG, Xu BW, Ni JQ. FlySAM transgenic CRISPRa system manual. Bio Protoc, 2019, 9(2):e3147.
doi: 10.21769/BioProtoc.3147 pmid: 33654892 |
| [40] |
Mao DC, Jia Y, Peng P, Shen D, Ren XJ, Zhu RB, Qiu YH, Han YT, Yu JC, Che QY, Li YT, Lu XY, Liu LP, Wang Z, Liu QF, Sun J, Ni JQ. Enhanced efficiency of flySAM by optimization of sgRNA parameters in Drosophila. G3 (Bethesda), 2020, 10(12):4483-4488.
doi: 10.1534/g3.120.401614 |
| [1] | Zhong Bian, Dongping Cao, Wenshu Zhuang, Shuwei Zhang, Qiaoquan Liu, Lin Zhang. Revelation of rice molecular design breeding: the blend of tradition and modernity [J]. Hereditas(Beijing), 2023, 45(9): 718-740. |
| [2] | Bingzheng Wang, Chao Zhang, Jiali Zhang, Jin Sun. Conditional editing of the Drosophila melanogaster genome using single transcripts expressing Cas9 and sgRNA [J]. Hereditas(Beijing), 2023, 45(7): 593-601. |
| [3] | Junhao An, Xueying Zhao, Shouyi Qiao, Daru Lu, Yan Pi. Application of modern computer technology in classical genetics lab course——Development of a mobile, lightweight and high-precision batch identification system for genetic traits of Drosophila [J]. Hereditas(Beijing), 2023, 45(4): 354-363. |
| [4] | Chengxian Wang, Yikang S. Rong, Min Cui. The molecular mechanism of Drosophila restricting telomeric transposons [J]. Hereditas(Beijing), 2023, 45(3): 221-228. |
| [5] | Meizhen Liu, Liren Wang, Yongmei Li, Xueyun Ma, Honghui Han, Dali Li. Generation of genetically modified rat models via the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2023, 45(1): 78-87. |
| [6] | Siyuan Xu, Jia Shou, Qiang Wu. Additional evidence of HS5-1 enhancer eRNA PEARL for protocadherin alpha gene regulation [J]. Hereditas(Beijing), 2022, 44(8): 695-764. |
| [7] | Xiaojun Zhang, Kun Xu, Juncen Shen, Lu Mu, Hongrun Qian, Jieyu Cui, Baoxia Ma, Zhilong Chen, Zhiying Zhang, Zehui Wei. A CRISPR/Cas9-Gal4BD donor adapting system for enhancing homology-directed repair [J]. Hereditas(Beijing), 2022, 44(8): 708-719. |
| [8] | Chong Zhang, Zixuan Wei, Min Wang, Yaosheng Chen, Zuyong He. Editing MC1R in human melanoma cells by CRISPR/Cas9 and functional analysis [J]. Hereditas(Beijing), 2022, 44(7): 581-590. |
| [9] | Yao Liu, Xianhui Zhou, Shuhong Huang, Xiaolong Wang. Prime editing: a search and replace tool with versatile base changes [J]. Hereditas(Beijing), 2022, 44(11): 993-1008. |
| [10] | Guangwu Yang, Yuan Tian. The F-box gene Ppa promotes lipid storage in Drosophila [J]. Hereditas(Beijing), 2021, 43(6): 615-622. |
| [11] | Hongyan Lin, Xuan Wang, Cong He, Ziling Zhou, Minkai Yang, Zhongling Wen, Hongwei Han, Guihua Lu, Jinliang Qi, Yonghua Yang. Progress on biosynthesis and function of the natural products of Zi Cao as a traditional Chinese medicinal herb [J]. Hereditas(Beijing), 2021, 43(5): 459-472. |
| [12] | Dingwei Peng, Ruiqiang Li, Wu Zeng, Min Wang, Xuan Shi, Jianhua Zeng, Xiaohong Liu, Yaoshen Chen, Zuyong He. Editing the cystine knot motif of MSTN enhances muscle development of Liang Guang Small Spotted pigs [J]. Hereditas(Beijing), 2021, 43(3): 261-270. |
| [13] | Xuewen Liu, Hongmei Wu, Ying Bai, Qun Zeng, Zemin Cao, Xiushan Wu, Min Tang. Potassium channel Shaker play a protective role against cardiac aging in Drosophila [J]. Hereditas(Beijing), 2021, 43(1): 94-99. |
| [14] | Na Wang, Zhilian Jia, Qiang Wu. RFX5 regulates gene expression of the Pcdhα cluster [J]. Hereditas(Beijing), 2020, 42(8): 760-774. |
| [15] | Guoling Li, Shanxin Yang, Zhenfang Wu, Xianwei Zhang. Recent developments in enhancing the efficiency of CRISPR/Cas9- mediated knock-in in animals [J]. Hereditas(Beijing), 2020, 42(7): 641-656. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||