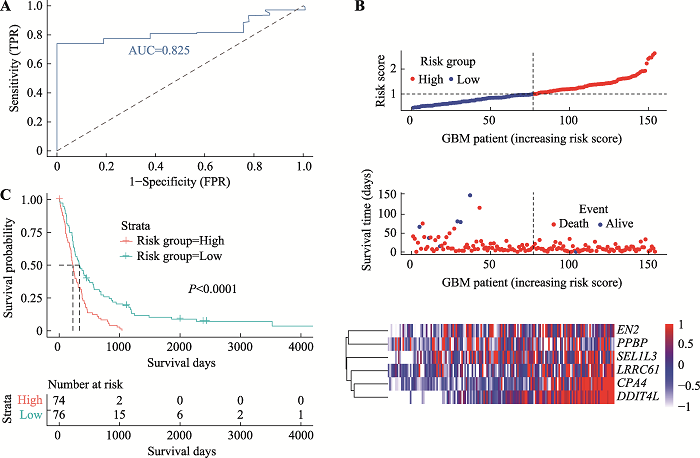
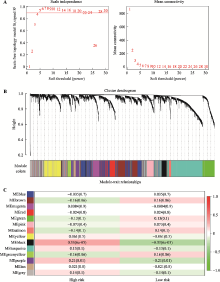
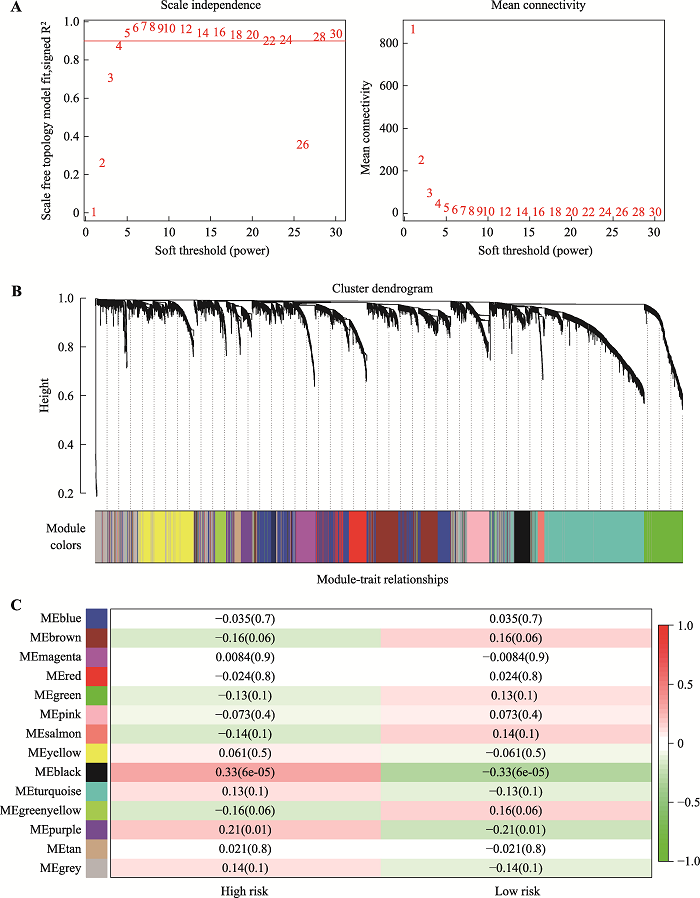
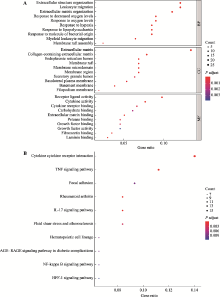
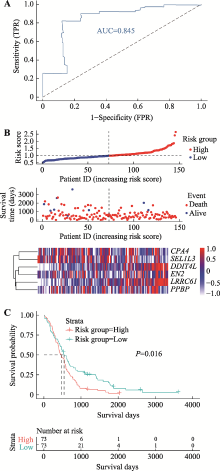
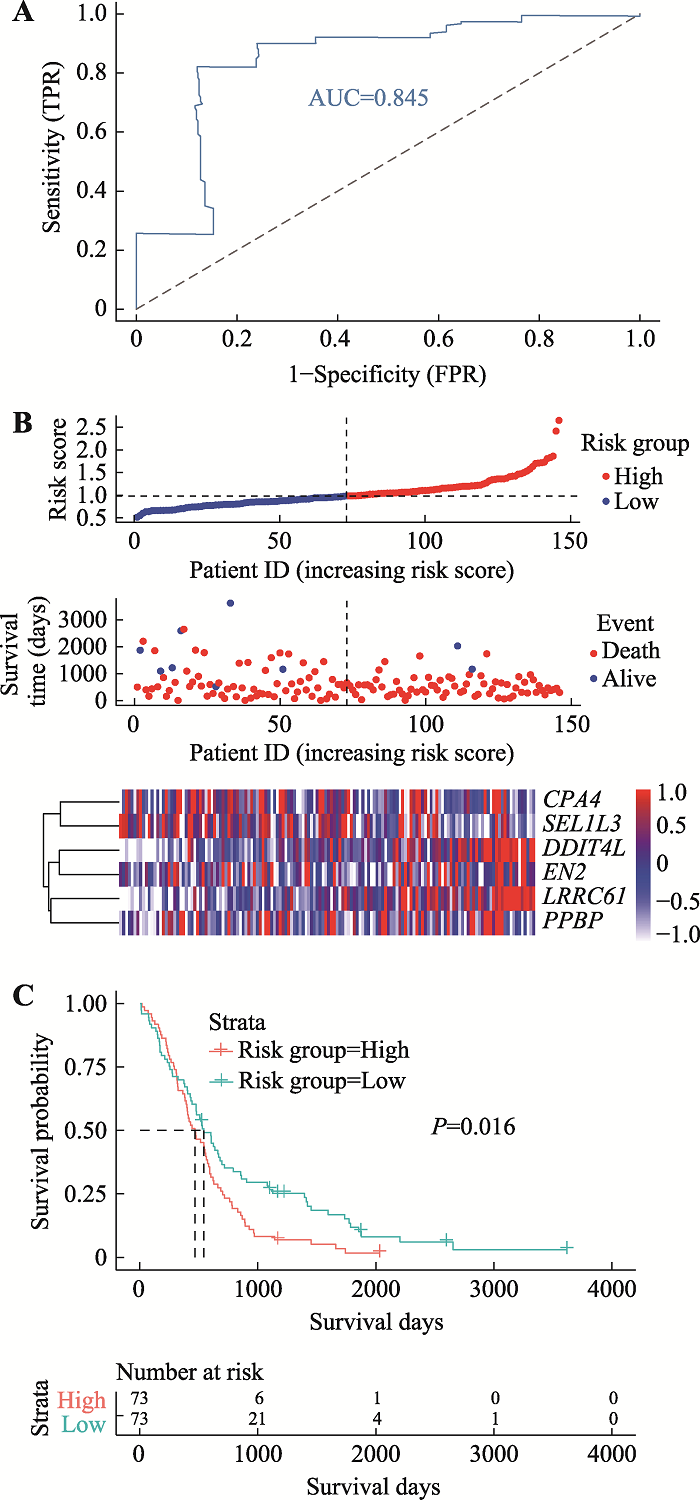
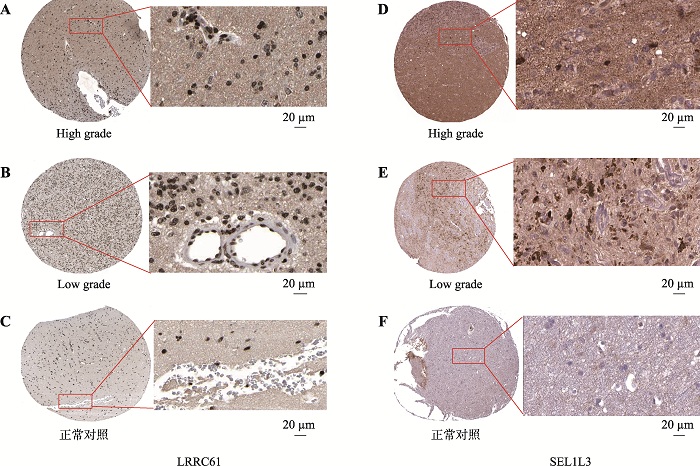
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (7): 665-679.doi: 10.16288/j.yczz.20-428
• Research Article • Previous Articles Next Articles
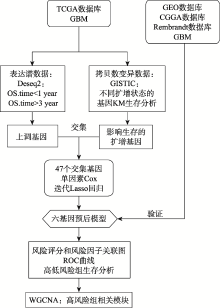
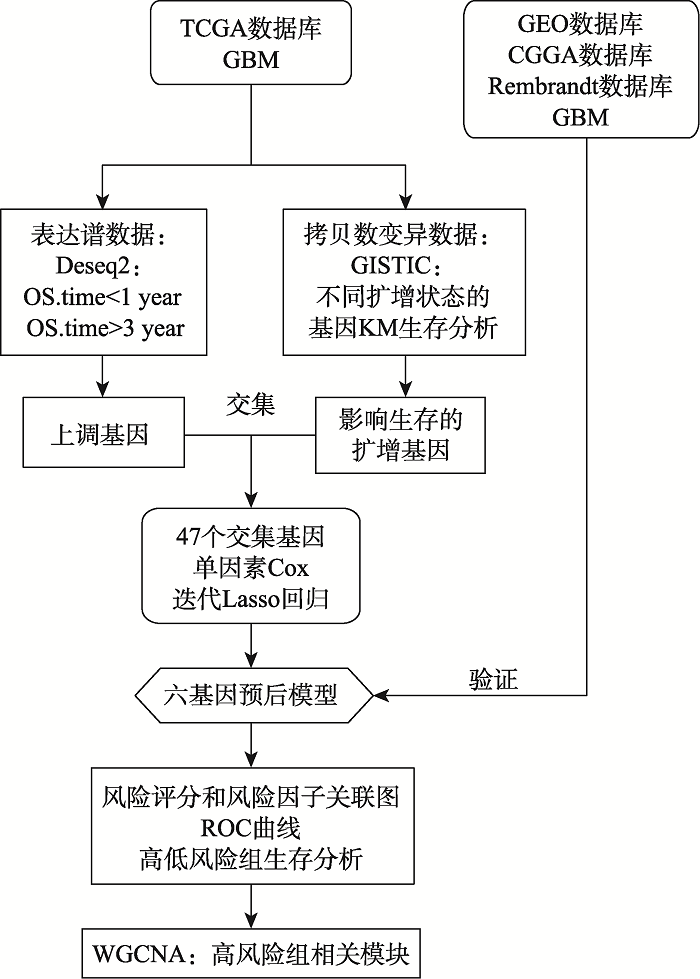
Establish six-gene prognostic model for glioblastoma based on multi-omics data of TCGA database
Changgui Lei( ), Xueyuan Jia(
), Xueyuan Jia( ), Wenjing Sun(
), Wenjing Sun( )
)
- Laboratory of Medical Genetics, Harbin Medical University, Key Laboratory of Preservation of Human Genetic Resources and Disease Control in China (Harbin Medical University), Ministry of Education, Harbin 150081, China
-
Received:2020-12-11Revised:2021-03-20Online:2021-07-20Published:2021-04-07 -
Contact:Sun Wenjing E-mail:leichanggui@hrbmu.edu.cn;jiaxueyuan@hrbmu.edu.cn;sunwj@ems.hrbmu.edu.cn -
Supported by:Supported by Program for Changjiang Scholars and Innovative Research Team in University No(IRT1230)
Cite this article
Changgui Lei, Xueyuan Jia, Wenjing Sun. Establish six-gene prognostic model for glioblastoma based on multi-omics data of TCGA database[J]. Hereditas(Beijing), 2021, 43(7): 665-679.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
"
| 基因 | HR | 95%CI | P值 | Log2(fold change) |
|---|---|---|---|---|
| COL1A2 | 1.209 | 0.972~1.502 | 0.088 | 1.686 |
| CPA4 | 1.268 | 1.018~1.579 | 0.034 | 1.238 |
| CXCL5 | 1.343 | 1.070~1.686 | 0.011 | 3.125 |
| CXCL6 | 1.498 | 1.006~2.231 | 0.047 | 2.908 |
| DDIT4L | 1.416 | 1.137~1.764 | 0.002 | 1.274 |
| EN2 | 1.716 | 1.290~2.282 | 0 | 1.840 |
| FLNC | 1.282 | 1.062~1.546 | 0.010 | 1.339 |
| LAMB1 | 1.227 | 0.981~1.534 | 0.073 | 1.232 |
| LRRC61 | 1.432 | 1.168~1.756 | 0.001 | 1.852 |
| MXRA8 | 1.298 | 1.047~1.608 | 0.017 | 1.113 |
| MYO1G | 1.236 | 0.995~1.534 | 0.055 | 1.242 |
| NPTX2 | 1.174 | 0.976~1.412 | 0.088 | 1.492 |
| PDLIM1 | 1.246 | 1.002~1.548 | 0.048 | 1.003 |
| PPBP | 1.284 | 0.968~1.702 | 0.083 | 2.439 |
| SDK1 | 1.178 | 0.971~1.428 | 0.096 | 1.088 |
| SEL1L3 | 1.317 | 1.098~1.580 | 0.003 | 1.560 |
| SOD3 | 1.234 | 1.021~1.490 | 0.029 | 1.049 |
| VGF | 1.322 | 1.074~1.627 | 0.008 | 1.884 |
Table 2
Multivariate Cox regression proportional hazard model"
| 数据集 | 变量 | HR | 95%CI | P值 | |
|---|---|---|---|---|---|
| TCGA GBM实验组 | 低风险组 | 1(ref) | 1(ref) | ||
| 高风险组 | 2.39 | 1.582~3.617 | 0.0004 | ||
| 年龄 | 1.02 | 1.005~1.04 | 0.01056 | ||
| 性别(女) | 1(ref) | 1(ref) | |||
| 性别(男) | 0.83 | 0.557~1.256 | 0.38845 | ||
| IDH野生型 | 1(ref) | 1(ref) | |||
| IDH突变型 | 0.69 | 0.202~2.357 | 0.55348 | ||
| GEO GSE16011验证组 | 低风险组 | 1(ref) | 1(ref) | ||
| 高风险组 | 1.46 | 1.012~2.125 | 0.04285 | ||
| 年龄 | 1.03 | 1.02~1.052 | 0.0001 | ||
| CGGA验证组 | 低风险组 | 1(ref) | 1(ref) | ||
| 高风险组 | 1.92 | 1.189~3.102 | 0.00766 | ||
| 年龄 | 1.01 | 0.989~1.028 | 0.38646 | ||
| Rembrandt验证组 | 低风险组 | 1(ref) | 1(ref) | ||
| 高风险组 | 1.76 | 1.18~2.61 | 0.005 | ||
| 性别(女) | 1(ref) | 1(ref) | |||
| 性别(男) | 1.0058 | 0.68~1.49 | 0.977 | ||
Supplementary Table 1
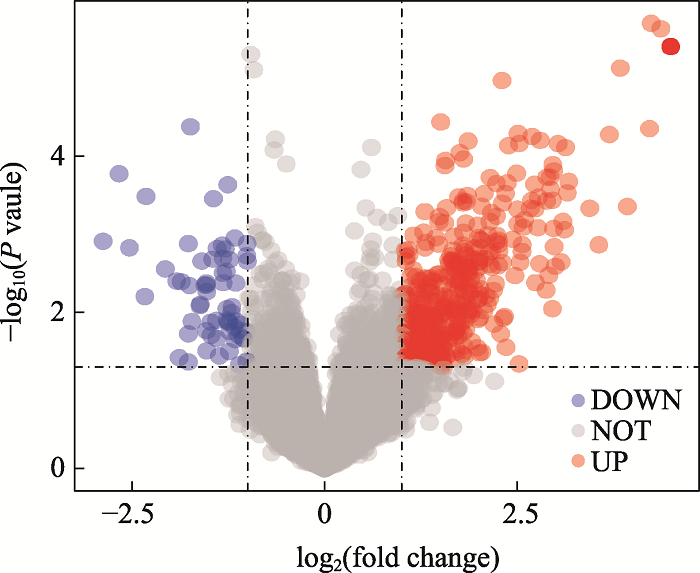
Differential genes of GBM patients with different survival time in TCGA"
| 基因 | Log2(fold change) | P值 | Diff type | 基因 | Log2(fold change) | P值 | Diff type |
|---|---|---|---|---|---|---|---|
| PCDHGB4 | -4.56 | 2.46e-21 | DOWN | CXCL14 | 2.53 | 6.87e-05 | UP |
| SAA1 | 4.24 | 1.99e-06 | UP | MCTP2 | 3.03 | 6.89e-05 | UP |
| SAA2 | 4.36 | 2.32e-06 | UP | COL5A1 | 2.39 | 7.29e-05 | UP |
| PI3 | 4.49 | 3.95e-06 | UP | CXCL5 | 3.13 | 7.71e-05 | UP |
| MARCO | 3.84 | 7.41e-06 | UP | SPAG4 | 1.75 | 8.98e-05 | UP |
| RPL39L | 2.30 | 1.07e-05 | UP | THBD | 1.80 | 0.000109 | UP |
| OSMR | 1.51 | 3.67e-05 | UP | NXPH4 | 1.57 | 0.000113 | UP |
| ZIC3 | -1.74 | 4.21e-05 | DOWN | RASSF9 | 2.96 | 0.000128 | UP |
| HOXB13 | 4.22 | 4.43e-05 | UP | SEL1L3 | 1.56 | 0.000132 | UP |
| KCNK5 | 2.51 | 5.12e-05 | UP | MMP1 | 2.97 | 0.000153 | UP |
| H19 | 3.70 | 5.28e-05 | UP | AQP9 | 2.49 | 0.000165 | UP |
| HOXC9 | 2.69 | 5.57e-05 | UP | REM1 | -2.67 | 0.000167 | DOWN |
| GPR1 | 2.80 | 6.26e-05 | UP | TMPRSS3 | 2.88 | 0.000185 | UP |
| CYGB | 1.86 | 6.39e-05 | UP | CLEC10A | 2.93 | 0.000185 | UP |
"
| 基因 | Log2(fold change) | P值 | Diff type | 基因 | Log2(fold change) | P值 | Diff type |
|---|---|---|---|---|---|---|---|
| HOXC6 | 2.15 | 0.000187 | UP | AQP5 | 1.99 | 0.000759 | UP |
| MMP7 | 3.17 | 0.00021 | UP | TFAP2C | 2.58 | 0.00081 | UP |
| PTX3 | 2.37 | 0.000219 | UP | CCL20 | 2.77 | 0.000822 | UP |
| RARRES1 | 2.23 | 0.000223 | UP | HOXC13 | 2.97 | 0.000842 | UP |
| COL1A1 | 2.72 | 0.000228 | UP | CCL18 | 3.11 | 0.000873 | UP |
| SPNS3 | -1.26 | 0.000231 | DOWN | MICAL2 | 1.29 | 0.000933 | UP |
| AREG | 2.93 | 0.000265 | UP | FBLN1 | 1.80 | 0.000938 | UP |
| PLA2G2A | 3.15 | 0.000294 | UP | CA9 | 2.12 | 0.00101 | UP |
| TREM1 | 2.07 | 0.000299 | UP | LIN7A | 1.16 | 0.00102 | UP |
| ULBP1 | 2.35 | 0.000299 | UP | SFRP5 | 2.24 | 0.00104 | UP |
| LRRC61 | 1.85 | 0.000317 | UP | LOXL1 | 1.68 | 0.00106 | UP |
| FLJ16779 | -2.32 | 0.000327 | DOWN | VWC2L | 1.99 | 0.0011 | UP |
| STC1 | 1.74 | 0.000332 | UP | NKX3-1 | 1.44 | 0.00111 | UP |
| CHODL | 2.77 | 0.000334 | UP | C4orf48 | -1.17 | 0.00111 | DOWN |
| GADD45G | -1.44 | 0.000349 | DOWN | PPBP | 2.44 | 0.00116 | UP |
| DLX5 | 2.96 | 0.000365 | UP | FCN3 | 2.02 | 0.00117 | UP |
| HOXB9 | 2.69 | 0.000366 | UP | RHOD | 2.25 | 0.00118 | UP |
| NNAT | 2.88 | 0.000373 | UP | ISLR2 | 2.03 | 0.00118 | UP |
| CCL2 | 1.79 | 0.000374 | UP | ZNF560 | -2.88 | 0.00122 | DOWN |
| EN2 | 1.84 | 0.000382 | UP | COL3A1 | 2.22 | 0.00123 | UP |
| BIRC3 | 1.80 | 0.000397 | UP | EMP2 | 1.32 | 0.00126 | UP |
| HOXC8 | 2.31 | 0.000398 | UP | DCTN3 | -1.01 | 0.00131 | DOWN |
| PTPRN | 2.04 | 0.000437 | UP | DCT | -1.77 | 0.00131 | DOWN |
| MAB21L2 | 3.93 | 0.00044 | UP | EEF1A2 | 1.48 | 0.00134 | UP |
| OLFM1 | 1.58 | 0.000456 | UP | TWIST2 | 2.26 | 0.00134 | UP |
| EBF3 | 2.67 | 0.000462 | UP | CD70 | 2.56 | 0.00135 | UP |
| CXCL13 | 3.44 | 0.000463 | UP | SLC18A3 | 3.56 | 0.00136 | UP |
| COL6A3 | 2.73 | 0.000469 | UP | IL1R1 | 1.79 | 0.00137 | UP |
| GATA3 | 2.50 | 0.000474 | UP | NOG | -1.33 | 0.00138 | DOWN |
| ULBP2 | 1.30 | 0.000511 | UP | FAM81B | 2.98 | 0.00143 | UP |
| ZFR2 | 2.16 | 0.000527 | UP | IL6 | 2.02 | 0.00147 | UP |
| SHISA6 | 2.23 | 0.000584 | UP | TSHR | -2.53 | 0.00148 | DOWN |
| ANK1 | 1.49 | 0.000589 | UP | NPR3 | 1.94 | 0.00148 | UP |
| ALDH1A3 | 2.59 | 0.000651 | UP | PLIN2 | 1.30 | 0.00149 | UP |
| EREG | 3.09 | 0.000683 | UP | CYP2E1 | -1.39 | 0.00151 | DOWN |
| IGFBP6 | 2.15 | 0.000683 | UP | CHI3L2 | 1.82 | 0.00151 | UP |
| SOCS3 | 1.48 | 0.000688 | UP | BMP2 | -1.29 | 0.00153 | DOWN |
| NFKBIZ | 1.56 | 0.000712 | UP | ITGA5 | 1.09 | 0.00156 | UP |
| ADAMTS2 | 1.83 | 0.000717 | UP | FAM20A | 1.66 | 0.00159 | UP |
| SFRP2 | 2.51 | 0.000734 | UP | SEMA3F | 1.04 | 0.00161 | UP |
"
| 基因 | Log2(fold change) | P值 | Diff type | 基因 | Log2(fold change) | P值 | Diff type |
|---|---|---|---|---|---|---|---|
| IBSP | 2.26 | 0.00162 | UP | CTHRC1 | 2.00 | 0.00263 | UP |
| MRC1 | 2.17 | 0.00165 | UP | CD300E | 2.17 | 0.00265 | UP |
| BARX1 | 2.56 | 0.00166 | UP | MXRA5 | 1.69 | 0.0027 | UP |
| VDR | 1.81 | 0.00168 | UP | GJB2 | 1.87 | 0.00274 | UP |
| CNPY1 | 2.14 | 0.00168 | UP | KCTD4 | -2.07 | 0.00277 | DOWN |
| TCTEX1D1 | 1.83 | 0.00183 | UP | RGS4 | 1.73 | 0.00282 | UP |
| P4HA2 | 1.10 | 0.00184 | UP | EMILIN2 | 1.15 | 0.00285 | UP |
| SDCBP2 | -1.01 | 0.00187 | DOWN | CPAMD8 | 1.70 | 0.00292 | UP |
| LOX | 1.64 | 0.00188 | UP | PLD4 | -1.28 | 0.00297 | DOWN |
| FSTL3 | 1.06 | 0.00192 | UP | TPBG | 1.95 | 0.00301 | UP |
| COCH | 1.88 | 0.00194 | UP | ADM2 | 1.81 | 0.00305 | UP |
| MYBPHL | -1.32 | 0.002 | DOWN | TSPAN31 | 1.86 | 0.00308 | UP |
| C9orf24 | 2.01 | 0.00201 | UP | S100A3 | -1.32 | 0.00313 | DOWN |
| SPAG17 | 2.08 | 0.00204 | UP | GRIN2A | 1.87 | 0.00314 | UP |
| TMEM100 | -1.45 | 0.00207 | DOWN | ADAMTS14 | 1.49 | 0.00317 | UP |
| COL12A1 | 1.94 | 0.0021 | UP | SVEP1 | 1.47 | 0.00323 | UP |
| RASL10A | -1.24 | 0.00211 | DOWN | CXCL6 | 2.91 | 0.00334 | UP |
| FAM110C | 1.61 | 0.00211 | UP | COL10A1 | 2.47 | 0.00337 | UP |
| MOCOS | 1.66 | 0.00214 | UP | RCAN2 | 1.60 | 0.00339 | UP |
| PDE4C | 1.65 | 0.00217 | UP | PLAUR | 1.12 | 0.00343 | UP |
| CSDC2 | -1.59 | 0.0022 | DOWN | SLC2A3 | 1.19 | 0.00347 | UP |
| KLK14 | -1.00 | 0.0022 | DOWN | ANGPTL4 | 1.53 | 0.00349 | UP |
| SLC6A6 | 1.06 | 0.0022 | UP | SH2D5 | 1.37 | 0.00359 | UP |
| LANCL2 | 2.00 | 0.00225 | UP | LTBP2 | 1.30 | 0.00361 | UP |
| CYP1B1 | 2.01 | 0.00225 | UP | FLNC | 1.34 | 0.00367 | UP |
| BMP5 | 3.07 | 0.00225 | UP | C5orf49 | 1.75 | 0.00371 | UP |
| FCGR2B | 1.67 | 0.00225 | UP | HS3ST3A1 | 1.89 | 0.00376 | UP |
| BTC | 2.54 | 0.00227 | UP | PTGES | 1.69 | 0.00378 | UP |
| CR1 | 2.31 | 0.00229 | UP | MMP9 | 1.97 | 0.00386 | UP |
| PCDHB4 | 1.62 | 0.00232 | UP | MYO1G | 1.24 | 0.00388 | UP |
| COL13A1 | 1.82 | 0.00233 | UP | CDH15 | -1.92 | 0.00393 | DOWN |
| TMEM233 | 1.74 | 0.00234 | UP | BICC1 | 1.40 | 0.00396 | UP |
| WIF1 | 2.79 | 0.00237 | UP | FBLN2 | 1.75 | 0.00397 | UP |
| PRRG3 | 2.14 | 0.00242 | UP | FGF17 | -1.86 | 0.00402 | DOWN |
| SLC16A10 | 1.70 | 0.00244 | UP | COL6A2 | 1.75 | 0.00409 | UP |
| IL7R | 1.74 | 0.00247 | UP | IGF2 | 1.79 | 0.00412 | UP |
| STMN2 | 2.26 | 0.0025 | UP | DKK1 | 2.74 | 0.00415 | UP |
| DLK1 | 2.99 | 0.00256 | UP | MMEL1 | -1.32 | 0.00415 | DOWN |
| S100A8 | 1.81 | 0.00256 | UP | CHST9 | -1.54 | 0.00419 | DOWN |
| THBS1 | 1.87 | 0.00261 | UP | TNFSF18 | -1.15 | 0.00421 | DOWN |
"
| 基因 | Log2(fold change) | P值 | Diff type | 基因 | Log2(fold change) | P值 | Diff type |
|---|---|---|---|---|---|---|---|
| LGR6 | 2.22 | 0.00428 | UP | THBS4 | 1.54 | 0.00678 | UP |
| COL1A2 | 1.69 | 0.00438 | UP | CAMK4 | 1.10 | 0.00686 | UP |
| ELF3 | 1.53 | 0.00439 | UP | RIT2 | 1.99 | 0.00689 | UP |
| RANBP17 | -1.56 | 0.00443 | DOWN | TPPP3 | 1.41 | 0.00709 | UP |
| NRXN3 | 1.33 | 0.00443 | UP | BDKRB2 | 1.46 | 0.00714 | UP |
| ACY3 | -1.53 | 0.00446 | DOWN | PCDHGB3 | 1.78 | 0.00719 | UP |
| RPRM | -1.76 | 0.00452 | DOWN | PDLIM1 | 1.00 | 0.00732 | UP |
| IL1R2 | 1.86 | 0.00462 | UP | LILRA5 | 1.71 | 0.00739 | UP |
| MXRA8 | 1.11 | 0.00466 | UP | CFH | 1.29 | 0.00744 | UP |
| GPX3 | 1.38 | 0.00477 | UP | CDK4 | 1.85 | 0.00746 | UP |
| RDH10 | 1.19 | 0.00478 | UP | LAMB1 | 1.23 | 0.00774 | UP |
| SLAMF8 | 1.36 | 0.00485 | UP | XKR7 | 2.11 | 0.00783 | UP |
| RPH3A | 2.01 | 0.00493 | UP | FERMT1 | -1.61 | 0.00784 | DOWN |
| ROR2 | 1.94 | 0.00497 | UP | MDM2 | 1.77 | 0.00793 | UP |
| TNFSF14 | 1.74 | 0.005 | UP | DHRS9 | -1.62 | 0.00816 | DOWN |
| ACTG2 | 1.32 | 0.00515 | UP | HSPG2 | 1.10 | 0.00818 | UP |
| CAPN6 | 2.88 | 0.00517 | UP | SLIT2 | 1.54 | 0.0082 | UP |
| F13A1 | 2.19 | 0.0052 | UP | DCHS2 | 1.32 | 0.00824 | UP |
| GALNT9 | 1.80 | 0.00527 | UP | CA14 | -1.20 | 0.00847 | DOWN |
| MAP3K9 | 1.19 | 0.00537 | UP | PRAME | 1.89 | 0.00857 | UP |
| CLEC9A | -1.53 | 0.00543 | DOWN | PDZK1IP1 | 1.53 | 0.00866 | UP |
| BHMT2 | 1.63 | 0.00549 | UP | MYL9 | 1.09 | 0.0087 | UP |
| MYBPH | 1.87 | 0.00552 | UP | TDO2 | 1.57 | 0.00879 | UP |
| EGFLAM | 1.38 | 0.00555 | UP | CHRDL2 | 1.69 | 0.00881 | UP |
| VGF | 1.88 | 0.00566 | UP | TRNP1 | 1.12 | 0.00888 | UP |
| VASN | 1.05 | 0.00571 | UP | CLEC3B | -1.27 | 0.00892 | DOWN |
| ICAM1 | 1.23 | 0.00574 | UP | MMP13 | 2.96 | 0.00894 | UP |
| FN1 | 1.04 | 0.00579 | UP | BNC2 | 1.47 | 0.00911 | UP |
| CD163 | 1.47 | 0.00588 | UP | SEZ6 | 1.67 | 0.00922 | UP |
| MAPK15 | 2.02 | 0.0059 | UP | GPC3 | 1.63 | 0.00923 | UP |
| TYW1B | 1.00 | 0.00602 | UP | SYT13 | 1.85 | 0.00931 | UP |
| CD209 | 2.09 | 0.00609 | UP | FAM183A | 1.92 | 0.00939 | UP |
| RIMBP2 | 1.55 | 0.00617 | UP | TBR1 | 1.69 | 0.00942 | UP |
| IGF2BP1 | 1.82 | 0.00621 | UP | IL2RA | 2.07 | 0.00944 | UP |
| DAPL1 | -2.34 | 0.00622 | DOWN | RNF128 | 1.88 | 0.00945 | UP |
| GPR68 | 1.30 | 0.00635 | UP | HTRA3 | 1.14 | 0.00947 | UP |
| HECW1 | 1.63 | 0.00651 | UP | SLC4A10 | 1.21 | 0.00994 | UP |
| NEFL | 2.14 | 0.0066 | UP | DCDC2 | 1.25 | 0.00996 | UP |
| GABRA4 | 1.99 | 0.00667 | UP | EN1 | 1.54 | 0.01 | UP |
| BCL11B | 1.26 | 0.00672 | UP | CX3CR1 | -1.21 | 0.01 | DOWN |
"
| 基因 | Log2(fold change) | P值 | Diff type | 基因 | Log2(fold change) | P值 | Diff type |
|---|---|---|---|---|---|---|---|
| CXCL1 | 1.67 | 0.0101 | UP | C1orf158 | 2.20 | 0.0141 | UP |
| CCDC8 | 1.27 | 0.0101 | UP | GALNT14 | -1.10 | 0.0142 | DOWN |
| GOLT1A | 1.85 | 0.0101 | UP | HAS1 | 1.47 | 0.0142 | UP |
| PPEF1 | 1.16 | 0.0104 | UP | CDCP1 | 1.09 | 0.0146 | UP |
| TNNT1 | 1.52 | 0.0105 | UP | MMP11 | 1.08 | 0.0147 | UP |
| HK3 | 1.30 | 0.0105 | UP | STAT4 | 1.18 | 0.0149 | UP |
| VSTM2A | 1.71 | 0.0106 | UP | EXPH5 | -1.17 | 0.0152 | DOWN |
| C1QL2 | 1.89 | 0.0107 | UP | FPR2 | 1.72 | 0.0153 | UP |
| TMEM26 | 1.07 | 0.0107 | UP | ME1 | 1.32 | 0.0154 | UP |
| PLA2R1 | 1.35 | 0.0107 | UP | ADAM8 | 1.25 | 0.0155 | UP |
| METTL1 | 1.36 | 0.0107 | UP | FNDC1 | 1.76 | 0.0155 | UP |
| PTPRU | 1.27 | 0.011 | UP | TNFAIP2 | 1.13 | 0.016 | UP |
| PCSK1 | 1.42 | 0.011 | UP | SP6 | 1.09 | 0.016 | UP |
| ANPEP | 1.40 | 0.0111 | UP | CD163L1 | 1.22 | 0.0162 | UP |
| DPT | 2.34 | 0.0113 | UP | HS3ST2 | 1.55 | 0.0163 | UP |
| PTGS2 | 1.48 | 0.0113 | UP | FAP | 1.38 | 0.0164 | UP |
| DOK2 | 1.29 | 0.0114 | UP | CHGA | 1.83 | 0.0166 | UP |
| EFCAB1 | 2.01 | 0.0117 | UP | FOSL1 | 1.16 | 0.0168 | UP |
| GSTM1 | 2.32 | 0.0118 | UP | ADAMTS15 | -1.10 | 0.0169 | DOWN |
| CABP1 | 1.34 | 0.012 | UP | PXDN | 1.11 | 0.017 | UP |
| FAM83G | 1.26 | 0.012 | UP | SOX3 | -1.54 | 0.0171 | DOWN |
| TIMP1 | 1.15 | 0.0121 | UP | ESM1 | 1.29 | 0.0171 | UP |
| MMP19 | 1.56 | 0.0122 | UP | GSX2 | 1.70 | 0.0173 | UP |
| LYVE1 | 1.99 | 0.0123 | UP | FBLIM1 | 1.04 | 0.0175 | UP |
| PNMT | 1.73 | 0.0124 | UP | SYN1 | 1.38 | 0.0175 | UP |
| TMPRSS9 | -1.27 | 0.0124 | DOWN | TNF | -1.23 | 0.0176 | DOWN |
| CPLX2 | 1.82 | 0.0124 | UP | GPC5 | 1.35 | 0.0176 | UP |
| AGAP2 | 1.69 | 0.0125 | UP | ADCYAP1R1 | -1.02 | 0.0177 | DOWN |
| RCN3 | 1.13 | 0.0125 | UP | TFPI2 | 1.66 | 0.0178 | UP |
| MLPH | 1.41 | 0.0125 | UP | MYT1L | 1.69 | 0.0178 | UP |
| ATP13A5 | -1.41 | 0.0125 | DOWN | C7 | 1.69 | 0.018 | UP |
| HYDIN | 1.43 | 0.0127 | UP | LRRC15 | 1.79 | 0.0182 | UP |
| CPS1 | -1.15 | 0.0128 | DOWN | MMP24 | 1.04 | 0.0182 | UP |
| MSTN | -1.72 | 0.013 | DOWN | OS9 | 1.07 | 0.0183 | UP |
| SOD3 | 1.05 | 0.0131 | UP | G0S2 | 1.11 | 0.0183 | UP |
| P2RY12 | -1.25 | 0.0132 | DOWN | DKK2 | 1.49 | 0.0186 | UP |
| C8orf34 | -1.48 | 0.0133 | DOWN | CSF3 | 2.28 | 0.0187 | UP |
| FOXJ1 | 1.46 | 0.0135 | UP | ZNF488 | -1.77 | 0.0187 | DOWN |
| C1QL1 | -1.24 | 0.0135 | DOWN | ELAVL2 | 1.56 | 0.0189 | UP |
| NTSR1 | 1.76 | 0.0137 | UP | MT1H | 1.43 | 0.0189 | UP |
"
| 基因 | Log2(fold change) | P值 | Diff type | 基因 | Log2(fold change) | P值 | Diff type |
|---|---|---|---|---|---|---|---|
| TSFM | 1.30 | 0.0191 | UP | SLC17A6 | 1.98 | 0.0283 | UP |
| ST6GALNAC1 | -1.12 | 0.0194 | DOWN | CTSK | 1.03 | 0.0283 | UP |
| SNX31 | -1.47 | 0.0195 | DOWN | GLRA2 | 1.54 | 0.029 | UP |
| CENPV | 1.21 | 0.0197 | UP | IL11 | 1.24 | 0.0291 | UP |
| CHIT1 | 1.49 | 0.0198 | UP | NPPA | 1.71 | 0.0295 | UP |
| CLIC6 | 1.74 | 0.0199 | UP | CHD5 | 1.37 | 0.0303 | UP |
| PCDHGB2 | 1.47 | 0.0208 | UP | TMEM130 | 1.28 | 0.0304 | UP |
| SNCB | 1.50 | 0.021 | UP | GABRD | 1.12 | 0.0308 | UP |
| ALPK2 | 1.50 | 0.0213 | UP | LUZP2 | -1.53 | 0.0309 | DOWN |
| NELL1 | 1.69 | 0.0214 | UP | PCDHB11 | 1.40 | 0.031 | UP |
| ENHO | -1.06 | 0.0215 | DOWN | ATP8A2 | 1.43 | 0.031 | UP |
| TMIGD2 | -1.40 | 0.0216 | DOWN | ITK | 1.17 | 0.0313 | UP |
| LRRC36 | -1.05 | 0.0219 | DOWN | NOS1 | 1.49 | 0.0314 | UP |
| SECTM1 | 1.05 | 0.0221 | UP | NEU4 | -1.24 | 0.0314 | DOWN |
| TAC3 | 1.51 | 0.0221 | UP | MME | 1.43 | 0.0315 | UP |
| CFTR | -1.16 | 0.0222 | DOWN | LOXL2 | 1.04 | 0.0316 | UP |
| HSPA7 | 1.14 | 0.0222 | UP | SDK1 | 1.09 | 0.0318 | UP |
| RSPO2 | 1.59 | 0.0224 | UP | PTPRR | 1.30 | 0.0319 | UP |
| EMILIN1 | 1.01 | 0.0225 | UP | UBE2QL1 | 1.15 | 0.0319 | UP |
| SH2D2A | 1.13 | 0.0229 | UP | GLRA3 | 2.04 | 0.032 | UP |
| SULT4A1 | 1.61 | 0.0231 | UP | TSHZ2 | 1.15 | 0.0326 | UP |
| DNAH2 | 1.24 | 0.0231 | UP | SERPINA5 | 1.10 | 0.0327 | UP |
| KRT17 | 1.41 | 0.0234 | UP | ARMC3 | 2.01 | 0.0328 | UP |
| SLC1A6 | 1.91 | 0.0234 | UP | GLT1D1 | 1.20 | 0.0328 | UP |
| PCDHB12 | 1.06 | 0.0238 | UP | OLFML1 | 1.07 | 0.033 | UP |
| SPON2 | 1.23 | 0.024 | UP | CPNE7 | 1.32 | 0.0332 | UP |
| TMEM200B | 1.21 | 0.0245 | UP | HPD | 1.23 | 0.0333 | UP |
| CLEC2B | 1.12 | 0.0247 | UP | WDR38 | 1.50 | 0.0335 | UP |
| ENTPD3 | 1.28 | 0.0254 | UP | DDIT4L | 1.27 | 0.0339 | UP |
| NPTX2 | 1.49 | 0.0259 | UP | MSX2 | 1.31 | 0.034 | UP |
| CCR2 | 1.45 | 0.0259 | UP | CPNE8 | 1.09 | 0.034 | UP |
| CNTN4 | 1.27 | 0.0267 | UP | HOTAIR | 1.77 | 0.034 | UP |
| NGFR | 1.10 | 0.0267 | UP | NDST3 | 1.79 | 0.0341 | UP |
| LRRIQ1 | 1.53 | 0.0268 | UP | NPTX1 | 1.37 | 0.0341 | UP |
| CPA4 | 1.24 | 0.0268 | UP | SLC6A15 | 1.32 | 0.0345 | UP |
| NEFM | 1.80 | 0.0268 | UP | ADAMDEC1 | 1.40 | 0.0349 | UP |
| AK7 | 1.36 | 0.0272 | UP | VSNL1 | 1.49 | 0.035 | UP |
| RGS7 | 1.12 | 0.0276 | UP | FABP5 | 1.08 | 0.035 | UP |
| MMP12 | 2.35 | 0.0279 | UP | LUM | 1.39 | 0.035 | UP |
| PCDHGA7 | 1.22 | 0.028 | UP | HTR1D | 1.38 | 0.0351 | UP |
"
| 基因 | Log2(fold change) | P值 | Diff type | 基因 | Log2(fold change) | P值 | Diff type |
|---|---|---|---|---|---|---|---|
| GABRA2 | 1.30 | 0.0353 | UP | NRGN | 1.15 | 0.0406 | UP |
| TCF23 | 1.58 | 0.0354 | UP | CHRNB2 | 1.05 | 0.041 | UP |
| ZBBX | -1.37 | 0.036 | DOWN | CLVS1 | 1.43 | 0.0412 | UP |
| CAMK2A | 1.37 | 0.0363 | UP | RAB3C | 1.35 | 0.0417 | UP |
| TP63 | 1.58 | 0.0364 | UP | OR51E1 | 1.13 | 0.0418 | UP |
| UBD | 1.43 | 0.0364 | UP | SCN3B | 1.02 | 0.0419 | UP |
| GATA4 | 1.81 | 0.0365 | UP | TFAP2A | 1.35 | 0.042 | UP |
| SYNGR3 | 1.11 | 0.0367 | UP | DACH1 | -1.01 | 0.0421 | DOWN |
| GNG3 | 1.32 | 0.0368 | UP | PODNL1 | 1.23 | 0.043 | UP |
| CELF4 | 1.34 | 0.0372 | UP | IGFN1 | -1.77 | 0.043 | DOWN |
| RYR3 | 1.19 | 0.0374 | UP | STEAP4 | 1.30 | 0.0432 | UP |
| BCL2A1 | 1.05 | 0.0375 | UP | CYP3A5 | 1.06 | 0.0437 | UP |
| ECEL1 | -1.89 | 0.0377 | DOWN | PCDHGA10 | 1.53 | 0.0438 | UP |
| OLFML2B | 1.04 | 0.0378 | UP | ISLR | 1.04 | 0.0439 | UP |
| SNAP25 | 1.40 | 0.0379 | UP | CLUL1 | 1.24 | 0.0441 | UP |
| CAMK1G | 1.42 | 0.0379 | UP | ST8SIA3 | 1.44 | 0.0448 | UP |
| IL18R1 | 1.17 | 0.038 | UP | SVOP | 1.44 | 0.045 | UP |
| FCN1 | 1.53 | 0.0381 | UP | IQSEC3 | 1.17 | 0.045 | UP |
| SERTAD4 | 1.27 | 0.0383 | UP | C4BPA | 2.52 | 0.0455 | UP |
| SHANK1 | 1.09 | 0.0383 | UP | ENPP5 | 1.27 | 0.0463 | UP |
| POPDC3 | 1.21 | 0.0385 | UP | CREG2 | 1.28 | 0.0464 | UP |
| SYT1 | 1.42 | 0.0386 | UP | C2CD4A | 1.47 | 0.0466 | UP |
| SLAMF6 | 1.08 | 0.0388 | UP | NKAIN4 | -1.11 | 0.0466 | DOWN |
| PCDHGB5 | 1.15 | 0.039 | UP | SDS | 1.02 | 0.0469 | UP |
| SP8 | 1.71 | 0.039 | UP | CHGB | 1.09 | 0.0474 | UP |
| GABRB2 | 1.47 | 0.0391 | UP | PRRX2 | 1.50 | 0.0481 | UP |
| IDO1 | 1.53 | 0.0394 | UP | LIF | 1.27 | 0.0491 | UP |
| WNT16 | 1.46 | 0.0397 | UP | FOSB | 1.54 | 0.0491 | UP |
| ICAM5 | 1.11 | 0.0402 | UP | MFAP2 | 1.04 | 0.0494 | UP |
| VGLL3 | 1.16 | 0.0402 | UP | KRT75 | 1.51 | 0.0497 | UP |
| PCOLCE2 | 1.14 | 0.0404 | UP | LYZ | 1.08 | 0.0497 | UP |
| HP | 1.46 | 0.0405 | UP |
Supplementary Table 2
The intersection of up-regulated genes in short survival group and SAG"
| 基因 | KM_P值 | Log2(fold change) | P值 | Diff type | 基因 | KM_P值 | Log2(fold change) | P值 | Diff type |
|---|---|---|---|---|---|---|---|---|---|
| AREG | 0.03 | 2.93 | 0.000265 | UP | LANCL2 | 0.01 | 2 | 0.00225 | UP |
| ARMC3 | 0.04 | 2.01 | 0.0328 | UP | LRRC61 | 0 | 1.85 | 0.000317 | UP |
| BTC | 0.03 | 2.54 | 0.00227 | UP | MAB21L2 | 0.03 | 3.93 | 0.00044 | UP |
| CNPY1 | 0.04 | 2.14 | 0.00168 | UP | MRC1 | 0.01 | 2.17 | 0.00165 | UP |
| COL1A2 | 0.02 | 1.69 | 0.00438 | UP | MXRA8 | 0.05 | 1.11 | 0.00466 | UP |
| CPA4 | 0.03 | 1.24 | 0.0268 | UP | MYO1G | 0.01 | 1.24 | 0.00388 | UP |
| CXCL1 | 0.02 | 1.67 | 0.0101 | UP | NDST3 | 0.01 | 1.79 | 0.0341 | UP |
| CXCL13 | 0.03 | 3.44 | 0.000463 | UP | NPTX2 | 0.02 | 1.49 | 0.0259 | UP |
| CXCL5 | 0.02 | 3.13 | 7.71e-05 | UP | PDLIM1 | 0.03 | 1 | 0.00732 | UP |
| CXCL6 | 0.02 | 2.91 | 0.00334 | UP | PPBP | 0.02 | 2.44 | 0.00116 | UP |
| CYP3A5 | 0.02 | 1.06 | 0.0437 | UP | SDK1 | 0 | 1.09 | 0.0318 | UP |
| DCHS2 | 0.03 | 1.32 | 0.00824 | UP | SEL1L3 | 0.05 | 1.56 | 0.000132 | UP |
| DDIT4L | 0.01 | 1.27 | 0.0339 | UP | SFRP2 | 0.03 | 2.51 | 0.000734 | UP |
| DLX5 | 0.03 | 2.96 | 0.000365 | UP | SLIT2 | 0 | 1.54 | 0.0082 | UP |
| EN2 | 0.04 | 1.84 | 0.000382 | UP | SOD3 | 0.01 | 1.05 | 0.0131 | UP |
| EREG | 0.03 | 3.09 | 0.000683 | UP | SPON2 | 0.03 | 1.23 | 0.024 | UP |
| FLNC | 0.04 | 1.34 | 0.00367 | UP | STEAP4 | 0.01 | 1.3 | 0.0432 | UP |
| GABRA2 | 0 | 1.3 | 0.0353 | UP | TDO2 | 0 | 1.57 | 0.00879 | UP |
| GABRA4 | 0 | 1.99 | 0.00667 | UP | TFPI2 | 0 | 1.66 | 0.0178 | UP |
| GABRD | 0 | 1.12 | 0.0308 | UP | TMEM130 | 0.02 | 1.28 | 0.0304 | UP |
| HECW1 | 0.01 | 1.63 | 0.00651 | UP | TYW1B | 0 | 1 | 0.00602 | UP |
| IL6 | 0.01 | 2.02 | 0.00147 | UP | VGF | 0.02 | 1.88 | 0.00566 | UP |
| IQSEC3 | 0.01 | 1.17 | 0.045 | UP | VSTM2A | 0 | 1.71 | 0.0106 | UP |
| LAMB1 | 0.04 | 1.23 | 0.00774 | UP |
| [1] |
Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol, 2006, 2(9):494-503.
pmid: 16932614 |
| [2] | Su CL, Li L, Chen XW, Zhang J, Shen NX, Wang ZX, Yang SQ, Li J, Zhu WZ, Wang CY. Summary of classification of central nervous system tumors in WHO 2016. Radiol Prac, 2016, 31(7):570-579. |
| 苏昌亮, 李丽, 陈小伟, 张巨, 申楠茜, 王振熊, 杨时骐, 李娟, 朱文珍, 王承缘. 2016年WHO中枢神经系统肿瘤分类总结. 放射学实践, 2016, 31(7):570-579. | |
| [3] |
Parsons DW, Jones S, Zhang XS, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SKN, Shinjo SMO, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science, 2008, 321(5897):1807-1812.
doi: 10.1126/science.1164382 |
| [4] |
Krex D, Klink B, Hartmann C, von Deimling A, Pietsch T, Simon M, Sabel M, Steinbach JP, Heese O, Reifenberger G, Weller M, Schackert G, German Glioma Network. Long-term survival with glioblastoma multiforme. Brain, 2007, 130:2596-2606.
doi: 10.1093/brain/awm204 |
| [5] |
Gerber NK, Goenka A, Turcan S, Reyngold M, Makarov V, Kannan K, Beal K, Omuro A, Yamada Y, Gutin P, Brennan CW, Huse JT, Chan TA. Transcriptional diversity of long-term glioblastoma survivors. Neuro Oncol, 2014, 16(9):1186-1195.
doi: 10.1093/neuonc/nou043 |
| [6] |
Asiedu MK, Thomas CF, Dong J, Schulte SC, Khadka P, Sun ZF, Kosari F, Jen J, Molina J, Vasmatzis G, Kuang R, Aubry MC, Yang P, Wigle DA. Pathways impacted by genomic alterations in pulmonary carcinoid tumors. Clin Cancer Res, 2018, 24(7):1691-1704.
doi: 10.1158/1078-0432.CCR-17-0252 |
| [7] |
Chang J, Tan WL, Ling ZQ, Xi RB, Shao MM, Chen MJ, Luo YY, Zhao YJ, Liu Y, Huang XC, Xia YC, Hu JL, Parker JS, Marron D, Cui QH, Peng LN, Chu JH, Li HM, Du ZL, Han YL, Tan W, Liu ZH, Zhan QM, Li Y, Mao WM, Wu C, Lin DX. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking- related mutation signature and genomic alterations. Nat Commun, 2017, 8:15290.
doi: 10.1038/ncomms15290 |
| [8] |
Liang L, Fang JY, Xu J. Gastric cancer and gene copy number variation: emerging cancer drivers for targeted therapy. Oncogene, 2016, 35(12):1475-1482.
doi: 10.1038/onc.2015.209 pmid: 26073079 |
| [9] | Wang Z, Wang LX. Gene amplification in lung cancer. Int J Genet, 2010, 33(3):155-159. |
| 王箴, 王良旭. 基因扩增在肺癌中的研究进展. 国际遗传学杂志, 2010, 33(3):155-159. | |
| [10] |
Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci, 2009, 100(12):2235-2241.
doi: 10.1111/cas.2009.100.issue-12 |
| [11] | Jin WX, Yang TM, Dong XY, Hua ZC, Xu XX. P53 mutation in human brain gliomas: Comparison of loss of heterozygosity with mutation frequency. Hereditas (Beijing), 2000, 22(6):357-360. |
| 金卫新, 杨天明, 董雪吟, 华子春, 徐贤秀. 脑胶质瘤中P53基因突变及其与染色体17p杂合性丢失的比较. 遗传, 2000, 22(6):357-360. | |
| [12] |
Wang YL, Zhao WJ, Xiao Z, Guan GF, Liu X, Zhuang MH. A risk signature with four autophagy-related genes for predicting survival of glioblastoma multiforme. J Cell Mol Med, 2020, 24(7):3807-3821.
doi: 10.1111/jcmm.v24.7 |
| [13] |
Lin WZ, Wu SH, Chen XC, Ye YL, Weng YL, Pan YH, Chen ZJ, Chen L, Qiu XX, Qiu SF. Characterization of hypoxia signature to evaluate the tumor immune microenvironment and predict prognosis in glioma groups. Front Oncol, 2020, 10:796.
doi: 10.3389/fonc.2020.00796 |
| [14] |
Wang JJ, Wang H, Zhu BL, Wang X, Qian YH, Xie L, Wang WJ, Zhu J, Chen XY, Wang JM, Ding ZL. Development of a prognostic model of glioma based on immune-related genes. Oncol Lett, 2021, 21(2):116.
doi: 10.3892/ol |
| [15] |
Hermansen SK, Sørensen MD, Hansen A, Knudsen S, Alvarado AG, Lathia JD, Kristensen BW. A 4-miRNA signature to predict survival in glioblastomas. PLoS One, 2017, 12(11):e0188090.
doi: 10.1371/journal.pone.0188090 |
| [16] | Jia DY, Lin W, Tang HL, Cheng YF, Xu KW, He YS, Geng WJ, Dai QX. Integrative analysis of DNA methylation and gene expression to identify key epigenetic genes in glioblastoma. Aging (Albany NY), 2019, 11(15):5579-5592. |
| [17] |
Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, Ceccarelli M, Bontempi G, Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res, 2016, 44(8):e71.
doi: 10.1093/nar/gkv1507 |
| [18] | Li X, Li MW, Zhang YN, Xu HM. Common cancer genetic analysis methods and application study based on TCGA database. Hereditas(Beijing), 2019, 41(3):234-242. |
| 李鑫, 李梦玮, 张依楠, 徐寒梅. 常用肿瘤基因分析方法及基于TCGA数据库的分析应用. 遗传, 2019, 41(3):234-242. | |
| [19] |
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014, 15(12):550.
doi: 10.1186/s13059-014-0550-8 |
| [20] |
Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du JY, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff I, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci USA, 2007, 104(50):20007-20012.
doi: 10.1073/pnas.0710052104 |
| [21] |
Stalpers LJA, Kaplan EL. Edward L. Kaplan and the Kaplan-Meier survival curve. BSHM Bulletin, 2018, 33(2):109-135.
doi: 10.1080/17498430.2018.1450055 |
| [22] |
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw, 2010, 33(1):1-22.
pmid: 20808728 |
| [23] |
Sveen A, Ågesen TH, Nesbakken A, Meling GI, Rognum TO, Liestøl K, Skotheim RI, Lothe RA. ColoGuidePro: a prognostic 7-gene expression signature for stage III colorectal cancer patients. Clin Cancer Res, 2012, 18(21):6001-6010.
doi: 10.1158/1078-0432.CCR-11-3302 |
| [24] |
Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med, 2013, 32(30):5381-5397.
doi: 10.1002/sim.5958 pmid: 24027076 |
| [25] |
Yu GC, Wang LG, Han YY, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 2012, 16(5):284-287.
doi: 10.1089/omi.2011.0118 |
| [26] | Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov M, Sergushichev A. Fast gene set enrichment analysis. bioRxiv, 2019: 060012. |
| [27] |
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 2008, 9:559.
doi: 10.1186/1471-2105-9-559 pmid: 19114008 |
| [28] | Zeng Y, Xiao HL, Guo QN. Research progress on the mechanism of epithelial mesenchymal transition in glioblastoma. Chin J Diagn Pathol, 2019, 26(8):533-538. |
| 曾英, 肖华亮, 郭乔楠. 胶质母细胞瘤上皮间叶转化机制研究进展. 诊断病理学杂志, 2019, 26(8):533-538. | |
| [29] |
Zeng S, Zhou C, Yang DH, Xu LS, Yang HJ, Xu MH, Wang H. LEF1-AS1 is implicated in the malignant development of glioblastoma via sponging miR-543 to upregulate EN2. Brain Res, 2020, 1736:146781.
doi: S0006-8993(20)30137-2 pmid: 32184164 |
| [30] |
Achyut BR, Shankar A, Iskander ASM, Ara R, Angara K, Zeng P, Knight RA, Scicli AG, Arbab AS. Bone marrow derived myeloid cells orchestrate antiangiogenic resistance in glioblastoma through coordinated molecular networks. Cancer Lett, 2015, 369(2):416-426.
doi: 10.1016/j.canlet.2015.09.004 pmid: 26404753 |
| [31] |
Du Q, Li EC, Liu YE, Xie WL, Huang C, Song JQ, Zhang W, Zheng YJ, Wang HL, Wang Q. CTAPIII/CXCL7: a novel biomarker for early diagnosis of lung cancer. Cancer Med, 2018, 7(2):325-335.
doi: 10.1002/cam4.2018.7.issue-2 |
| [32] |
Ross PL, Cheng I, Liu X, Cicek MS, Carroll PR, Casey G, Witte JS. Carboxypeptidase 4 gene variants and early-onset intermediate-to-high risk prostate cancer. BMC Cancer, 2009, 9:69.
doi: 10.1186/1471-2407-9-69 |
| [33] |
Pan HD, Pan JX, Ji L, Song SB, Lv H, Yang ZR, Guo YB. Carboxypeptidase A4 promotes cell growth via activating STAT3 and ERK signaling pathways and predicts a poor prognosis in colorectal cancer. Int J Biol Macromol, 2019, 138:125-134.
doi: 10.1016/j.ijbiomac.2019.07.028 |
| [34] | Simonson B, Subramanya V, Chan MC, Zhang AF, Franchino H, Ottaviano F, Mishra MK, Knight AC, Hunt D, Ghiran I, Khurana TS, Kontaridis MI, Rosenzweig A, Das S. DDiT4L promotes autophagy and inhibits pathological cardiac hypertrophy in response to stress. Sci Signal , 2017, 10(468): eaaf5967. |
| [1] | Min Cheng, Jing Zhang, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of the hypoxia-associated long non-coding RNA signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2022, 44(2): 153-167. |
| [2] | Hongbo Luo, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of a DNA methylation-driven transcriptional signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2020, 42(8): 775-787. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||