Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (3): 242-255.doi: 10.16288/j.yczz.23-210
• Research Article • Previous Articles Next Articles
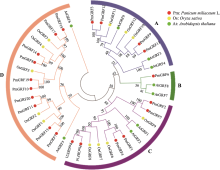
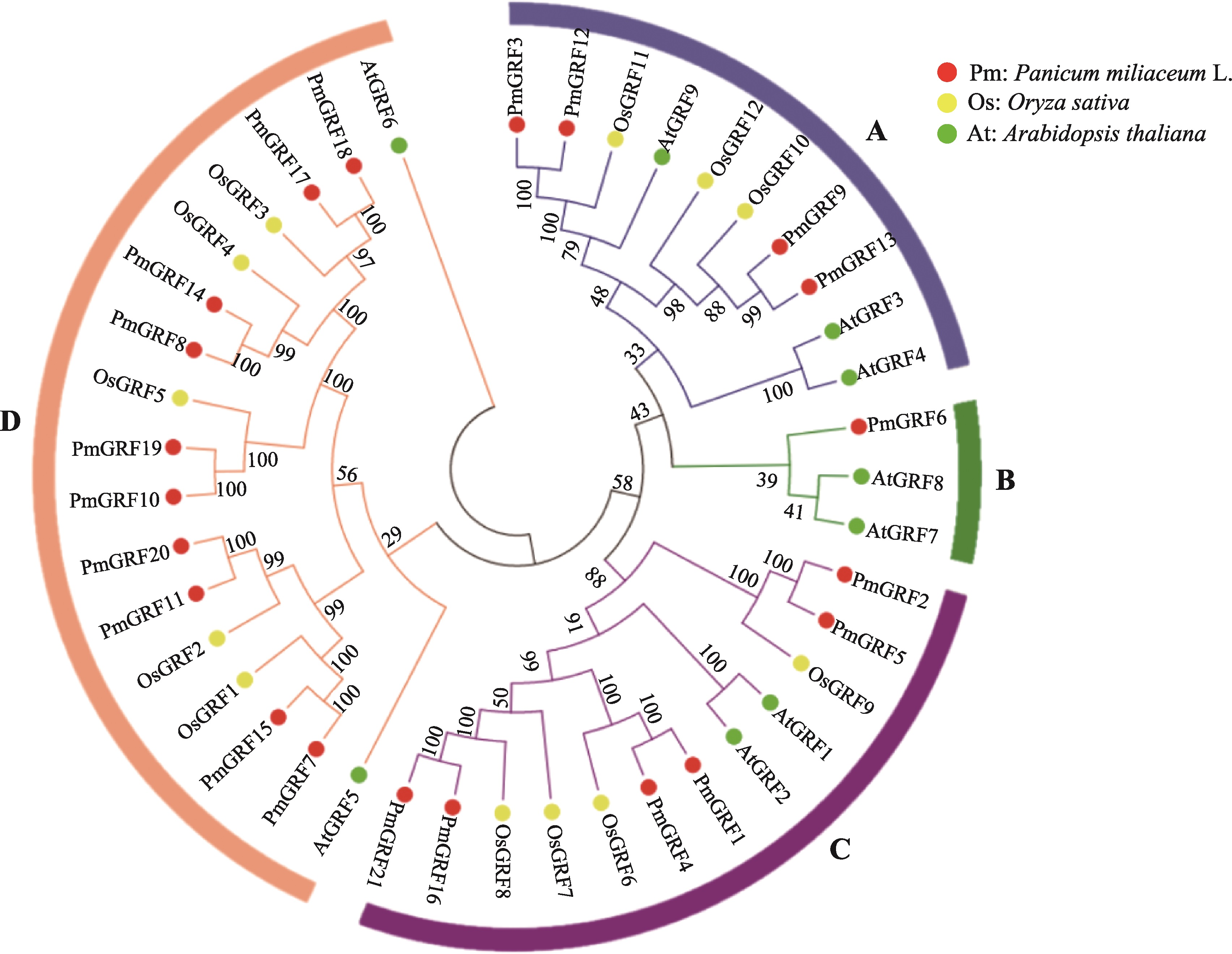
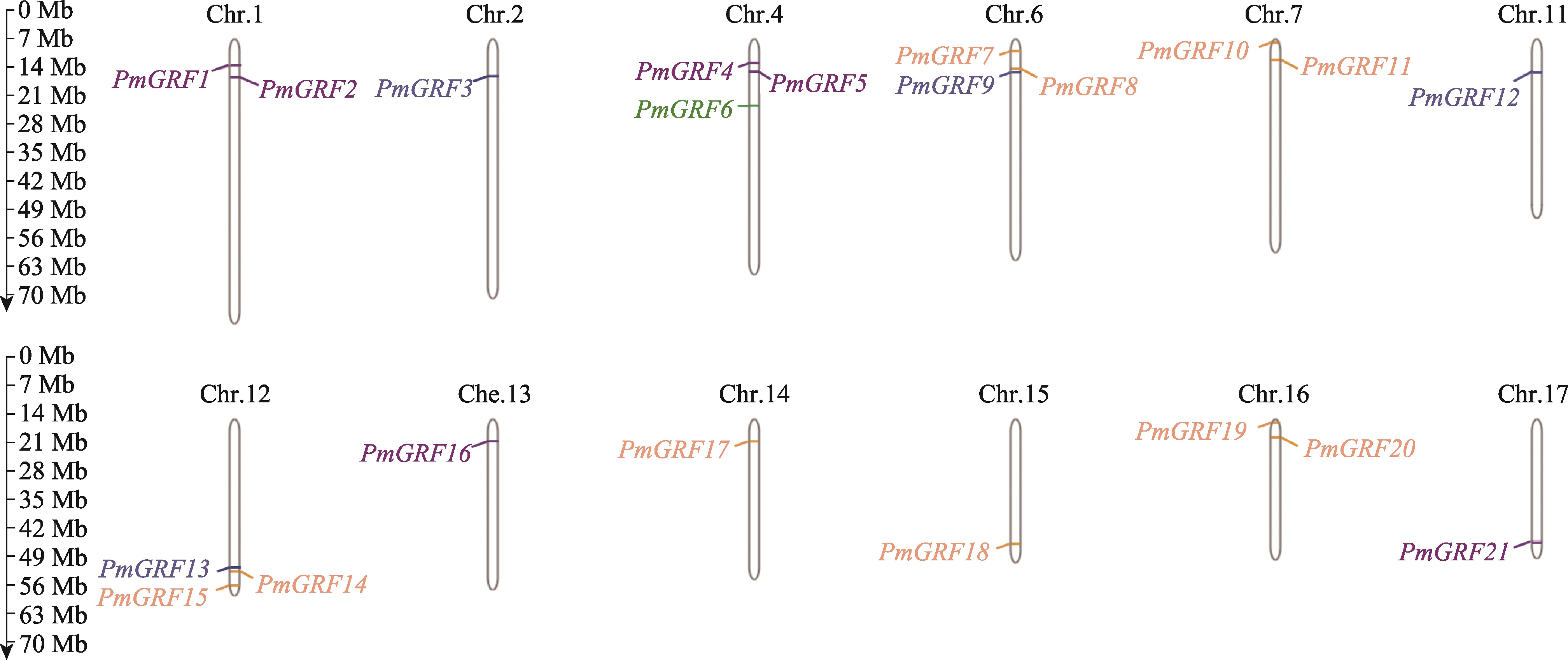
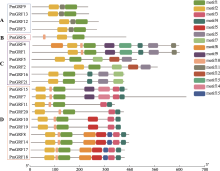
Genome-wide identification of GRF transcription factors and their expression profile in stem meristem of broomcorn millet (Panicum miliaceum L.)
Heng Wei1( ), Tianpeng Liu2, Jihong He2, Kongjun Dong2, Ruiyu Ren2, Lei Zhang2, Yawei Li2, Ziyi Hao1, Tianyu Yang1,2(
), Tianpeng Liu2, Jihong He2, Kongjun Dong2, Ruiyu Ren2, Lei Zhang2, Yawei Li2, Ziyi Hao1, Tianyu Yang1,2( )
)
- 1. College of Life Science and Technology, Gansu Agricultural University, Lanzhou 730070, China
2. Crop Research Institute, Gansu Academy of Agricultural Sciences, Lanzhou 730070, China
-
Received:2023-08-01Revised:2024-01-10Online:2024-03-20Published:2024-01-26 -
Contact:Tianyu Yang E-mail:1715414764@qq.com;13519638111@163.com -
Supported by:Second Tibetan Plateau Scientific Expedition and Research Program(2019QZKK0303);Science and Technology Planning Project of Gansu Province(22CX8NA028);China Modern Agricultural Industrial Technology System Project(CARS-06-14.5-A8)
Cite this article
Heng Wei, Tianpeng Liu, Jihong He, Kongjun Dong, Ruiyu Ren, Lei Zhang, Yawei Li, Ziyi Hao, Tianyu Yang. Genome-wide identification of GRF transcription factors and their expression profile in stem meristem of broomcorn millet (Panicum miliaceum L.)[J]. Hereditas(Beijing), 2024, 46(3): 242-255.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Sequences of the primers used for qRT-PCR"
| 基因名称 | 引物序列(5'→3') |
|---|---|
| PmGRF2 | F:GACACAGCACATGGCAACGA R:GTGAGCGGCACATCAACAGA |
| PmGRF3 | F:GCAGCAACAGCAGCACTTCA R:GACGAGATGAGTAGGCACAGGT |
| PmGRF4 | F:CGGTGCTGCTGGAGACTGAT R:GGCAACGATGTTCCGTGATTCT |
| PmGRF5 | F:TTGAGTCGTCACTGTTCTTGGA R:CGGACAACCTGCCTTACATCTA |
| PmGRF12 | F:GCAGCAACAGCAGCACTTCA R:GACGAGATGAGTAGGCACAGGT |
| PmGRF16 | F:GGTCGCAGTCCATTGGTCAGT R:CGGAGTAGGCAGTTCAGCAGAA |
| PmGRF21 | F:TGCCGTCCAGCTTGCTCCTT R:CACCGCCACTTCTTGCCATCAG |
| Pmactin | F:GGCATCACACCTTCTACAAC R:TCTCGAACATGATCTGGGTC |
Table 2
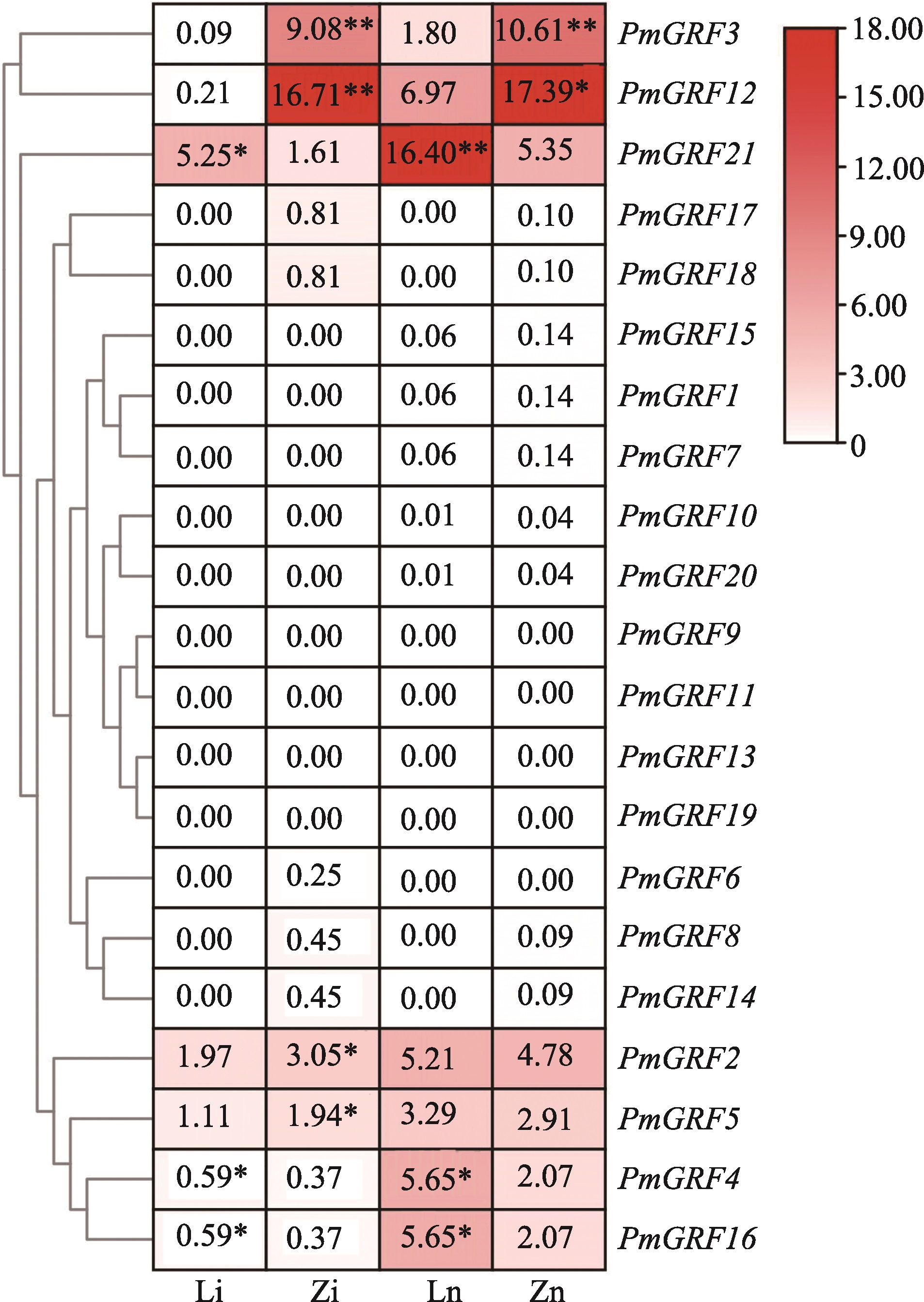
Physicochemical properties and subcellular localization in broomcorn millet GRF gene family"
| 基因名称 | 基因ID | 氨基酸数 | 分子质量 (Da) | 等电点 | 不稳定 指数 | 脂肪族氨基酸指数 | 亲水性平 均系数 | 亚细胞定位 | 基因染色 体定位 |
|---|---|---|---|---|---|---|---|---|---|
| PmGRF1 | LM01CHG000904 | 591 | 61,524.63 | 6.57 | 49.17 | 70.34 | -0.270 | 细胞核 | Chr.1 |
| PmGRF2 | LM01CHG001244 | 505 | 55,604.61 | 9.29 | 53.48 | 63.25 | -0.763 | 细胞核 | Chr.1 |
| PmGRF3 | LM02CHG001176 | 259 | 27,206.29 | 5.05 | 53.67 | 59.31 | -0.669 | 细胞核 | Chr.2 |
| PmGRF4 | LM04CHG000872 | 587 | 61,483.46 | 6.57 | 50.54 | 68.14 | -0.325 | 细胞核 | Chr.4 |
| PmGRF5 | LM04CHG001174 | 421 | 46,386.86 | 9.33 | 55.54 | 59.45 | -0.805 | 细胞核 | Chr.4 |
| PmGRF6 | LM04CHG002106 | 375 | 40,549.16 | 9.03 | 64.84 | 51.23 | -0.866 | 细胞核 | Chr.4 |
| PmGRF7 | LM06CHG000359 | 456 | 64,962.13 | 7.20 | 62.26 | 54.42 | -0.548 | 细胞核 | Chr.6 |
| PmGRF8 | LM06CHG000847 | 591 | 41,514.19 | 6.57 | 60.45 | 64.39 | -0.430 | 细胞核 | Chr.6 |
| PmGRF9 | LM06CHG000971 | 618 | 23,148.19 | 9.43 | 49.83 | 68.17 | -0.266 | 细胞核 | Chr.6 |
| PmGRF10 | LM07CHG000108 | 392 | 38,520.84 | 7.05 | 52.44 | 55.20 | -0.646 | 细胞核 | Chr.7 |
| PmGRF11 | LM07CHG000642 | 224 | 35,200.07 | 9.64 | 59.85 | 56.34 | -0.673 | 细胞核 | Chr.7 |
| PmGRF12 | LM11CHG001144 | 356 | 27,888.04 | 8.92 | 55.18 | 60.79 | -0.626 | 细胞核 | Chr.11 |
| PmGRF13 | LM12CHG002278 | 331 | 23,779.94 | 9.11 | 43.53 | 69.21 | -0.228 | 叶绿体、细胞核 | Chr.12 |
| PmGRF14 | LM12CHG002388 | 267 | 41,572.33 | 4.93 | 60.41 | 64.87 | -0.444 | 细胞核 | Chr.12 |
| PmGRF15 | LM12CHG002856 | 229 | 41,699.42 | 9.69 | 56.70 | 47.23 | -0.733 | 细胞核 | Chr.12 |
| PmGRF16 | LM13CHG000540 | 392 | 40,124.76 | 7.05 | 53.80 | 66.04 | -0.481 | 细胞核 | Chr.13 |
| PmGRF17 | LM14CHG000674 | 371 | 40,471.29 | 7.68 | 54.41 | 59.10 | -0.482 | 细胞核 | Chr.14 |
| PmGRF18 | LM15CHG002021 | 254 | 40,606.53 | 9.72 | 55.03 | 61.14 | -0.478 | 细胞核 | Chr.15 |
| PmGRF19 | LM16CHG000105 | 376 | 38,631.90 | 8.74 | 54.94 | 54.61 | -0.635 | 细胞核 | Chr.16 |
| PmGRF20 | LM16CHG000628 | 376 | 39,074.77 | 8.81 | 66.46 | 61.72 | -0.493 | 细胞核 | Chr.16 |
| PmGRF21 | LM17CHG001185 | 358 | 40,177.86 | 8.93 | 53.50 | 66.46 | -0.466 | 细胞核 | Chr.17 |
| [1] | Liebsch D, Palatnik JF. MicroRNA miR396, GRF transcription factors and GIF co-regulators: a conserved plant growth regulatory module with potential for breeding and biotechnology. Curr Opin Plant Bioly, 2020, 53: 31-42. |
| [2] | Yang XR, He SE, Chen SX. Research progress of GRF transcription factors in plants. Eucalyptus Sci & Technol, 2022, 39(3): 57-66. |
| 杨雪芮, 何沙娥, 陈少雄. GRF转录因子在植物中的研究进展. 桉树科技, 2022, 39(3): 57-66. | |
| [3] |
Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell, 2004, 119(1): 109-120.
doi: 10.1016/j.cell.2004.09.018 |
| [4] | Shi PB, He B, Fei YY, Wang J, Wang WY, Wei FY, Lv YD, Gu MF. Identification and expression analysis of GRF transcription factor family of Chenopodium quinoa. Acta Agron Sin, 2019, 45(12): 1841-1850. |
|
时丕彪, 何冰, 费月跃, 王军, 王伟义, 魏福友, 呂远大, 顾闽峰. 藜麦GRF转录因子家族的鉴定及表达分析. 作物学报, 2019, 45(12): 1841-1850.
doi: 10.3724/SP.J.1006.2019.94049 |
|
| [5] |
Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci USA, 2004, 101(36): 13374-13379.
doi: 10.1073/pnas.0405450101 |
| [6] |
Rodriguez RE, Ercoli MF, Debernardi JM, Breakfield NW, Mecchia MA, Sabatini M, Cools T, de Veyldeer L, Benfey PN, Palatnik JF. MicroRNA miR396 regulates the switch between stem cells and transit-amplifying cells in Arabidopsis roots. Plant Cell, 2015, 27(12): 3354-3366.
doi: 10.1105/tpc.15.00452 |
| [7] | Dai MY, Gao M, Li WC. Bioinformatics identification and expression analysis of GRF transcription factor family of Castor Bean. Mol Plant Breed, 2019, 19(22): 7383-7390. |
| 代梦媛, 高梅, 李文昌. 蓖麻GRF转录因子家族生物信息学鉴定及表达分析. 分子植物育种, 2021, 19(22): 7383-7390. | |
| [8] |
Huang WD, He YQ, Yang L, Lu C, Zhu YX, Sun C, Ma DF, Yin JL. Genome-wide analysis of growth-regulating factors (GRFs) in Triticum aestivum. PeerJ, 2021, 9: e10701.
doi: 10.7717/peerj.10701 |
| [9] | Yuan Q, Zhang CL, Zhao TT, Xu XY. Research advances of GRF transcription factor in plant. Genomics Appl Biol, 2017, 36(8): 3145-3151. |
| 袁岐, 张春利, 赵婷婷, 许向阳. 植物中GRF转录因子的研究进展. 基因组学与应用生物学, 2017, 36(8): 3145-3151. | |
| [10] |
van der Knaap E, Kim JH, Kende H. A novel gibberellin- induced gene from rice and its potential regulatory role in stem growth. Plant Physiol, 2000, 122(3): 695-704.
doi: 10.1104/pp.122.3.695 pmid: 10712532 |
| [11] |
Choi D, Kim JH, Kende H. Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol, 2004, 45(7): 897-904.
doi: 10.1093/pcp/pch098 |
| [12] |
Kim JH, Choi D, Kende H. The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J, 2003, 36(1): 94-104.
pmid: 12974814 |
| [13] |
Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MACJ, de Vries SC. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell, 2003, 15(7): 1563-1577.
doi: 10.1105/tpc.012203 pmid: 12837947 |
| [14] |
Chen HL, Ge WN. Identification, molecular characteristics, and evolution of GRF gene family in foxtail millet (Setaria italica L.). Front Genet, 2022, 12: 727674.
doi: 10.3389/fgene.2021.727674 |
| [15] |
Zhang DF, Li B, Jia GQ, Zhang TF, Dai JR, Li JS, Wang SC. Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in maize (Zea mays L.). Plant Sci, 2008, 175(6): 809-817.
doi: 10.1016/j.plantsci.2008.08.002 |
| [16] |
Chen F, Yang YZ, Luo XF, Zhou WG, Dai YJ, Zheng C, Liu WG, Yang WY, Shu K. Genome-wide identification of GRF transcription factors in soybean and expression analysis of GmGRF family under shade stress. BMC Plant Biol, 2019, 19(1): 269.
doi: 10.1186/s12870-019-1861-4 |
| [17] |
Zhang JF, Li ZF, Jin JJ, Xie XD, Zhang H, Chen QS, Luo ZP, Yang J. Genome-wide identification and analysis of the growth-regulating factor family in tobacco (Nicotiana tabacum). Gene, 2018, 639: 117-127.
doi: S0378-1119(17)30804-1 pmid: 28978430 |
| [18] | Wang Y, Zhang LN, Tang FY, Zhao XX, Xi YJ, Wang WW. Genome-wide identification and analysis of GRF transcription factors family in switchgrass. Acta Agrestia Sinica, 2019, 30(3): 575-586. |
|
王燕, 张礼宁, 唐方毅, 赵晓晓, 奚亚军, 王伟伟. 柳枝稷GRF转录因子家族全基因组鉴定与分析. 草地学报, 2022, 30(3): 575-586.
doi: 10.11733/j.issn.1007-0435.2022.03.009 |
|
| [19] | Jin L, Hass Agula, Gao F. Genome-wide identification and analysis of growth regulating factor genes(GRF) in cucumis melo L. Genomics Appl Biol, 2020, 39(8): 3554-3560. |
| 金兰, 哈斯阿古拉, 高峰. 甜瓜GRF转录因子的全基因组鉴定和分析. 基因组学与应用生物学, 2020, 39(8): 3554-3560. | |
| [20] |
Liu L, Li XJ, Li B, Sun MY, Li SX. Genome-wide analysis of the GRF gene family and their expression profiling in peach (Prunus persica). J Plant Interact, 2022, 17(1): 437-449.
doi: 10.1080/17429145.2022.2045370 |
| [21] | Li ZQ, Xie Q, Yan JH, Chen JQ, Chen QX. Genome-wide identification and characterization of the abiotic-stress- responsive GRF gene family in diploid woodland strawberry (Fragaria vesca). Plants (Basel), 2021, 10(9): 1916. |
| [22] |
Huang J, Chen GZ, Ahmad S, Hao Y, Chen JL, Zhou YZ, Lan SR, Liu ZJ, Peng DH. Genome-wide identification and characterization of the GRF gene family in Melastoma dodecandrum. Int J Mol Sci, 2023, 24(2): 1261.
doi: 10.3390/ijms24021261 |
| [23] |
Dong KJ, Liu TP, He JH, Ren RY, Zhang L, Yang TY. Evaluation and identification indexes selection on the drought resistance of broomcorn millet bred cultivars at seeding stage. J Plant Genet Resour, 2015, 16(5): 968-975.
doi: 10.13430/j.cnki.jpgr.2015.05.007 |
|
董孔军, 刘天鹏, 何继红, 任瑞玉, 张磊, 杨天育. 糜子育成品种苗期抗旱性评价与鉴定指标筛选. 植物遗传资源学报, 2015, 16(5): 968-975.
doi: 10.13430/j.cnki.jpgr.2015.05.007 |
|
| [24] |
Yuan YH, Liu CJ, Gao YB, Ma Q, Yang QH, Feng BL. Proso millet (Panicum miliaceum L.): a potential crop to meet demand scenario for sustainable saline agriculture. J Environ Manage, 2021, 296: 113216.
doi: 10.1016/j.jenvman.2021.113216 |
| [25] |
Yang P, Panhwar RB, Li J, Gao JF, Gao XL, Wang PK, Feng BL. Changes of yield and traits of broomcorn millet cultivars in China based on the data from national cultivars regional adaptation test. Sci Agric Sin, 2017, 50(23):4517-4529.
doi: 10.3864/j.issn.0578-1752.2017.23.006 |
|
杨璞, Rabia Begum Panhwar, 李境, 高金锋, 高小丽, 王鹏科, 冯佰利. 基于国家品种区域试验数据的中国糜子品种产量和性状变化. 中国农业科学, 2017, 50(23): 4517-4529.
doi: 10.3864/j.issn.0578-1752.2017.23.006 |
|
| [26] |
Lin FY, Wang SQ, Hu YG, He BR.Cloning of A S-adenosylmethionine synthetase gene from broomcorn millet (Panicum miliaceum L.) and its expression during drought and re-watering. Acta Agron Sin, 2008, 34(5): 777-782.
doi: 10.3724/SP.J.1006.2008.00777 |
| 林凡云, 王士强, 胡银岗, 何蓓如. 糜子SAMS基因的克隆及其在干旱复水中的表达模式分析. 作物学报, 2008, 34(5): 777-782. | |
| [27] |
Lin FY, Wang SQ, Hu YG, He BR. Cloning and expression analysis of drought-tolerant and water saving gene PmMYB in broomcorn millet. Hereditas (Beijing), 2008, 30(3): 373-379.
doi: 10.3724/SP.J.1005.2008.00373 |
| 林凡云, 王士强, 胡银岗, 何蓓如. 糜子抗旱节水相关基因PmMYB的克隆及表达分析. 遗传, 2008, 30(3): 373-379. | |
| [28] | Pan WX, Liu TP, He JH, Dong KJ, Ren RY, Zhang L, Yang TY. Genome-wide identification and expression characteristics of the YABBY gene family under hypertonic solution stress in broomcorn millet (Panicum miliaceum L.). Genomics Appl Biol, 2022, 41(5): 1067-1078. |
| 盘婉向, 刘天鹏, 何继红, 董孔军, 任瑞玉, 张磊, 杨天育. 糜子(Panicum miliaceum L.)全基因组YABBY基因家族鉴定与高渗溶液胁迫下表达特征. 基因组学与应用生物学, 2022, 41(5): 1067-1078. | |
| [29] | Wang M, Liu TP, He JH, Dong KJ, Ren RY, Zhang L, Yang TY. Genome-wide identification of bZIP gene family in broomcorn millet and analysis of its expression characteristics under polyethylene glycol treatment in seedling stage. Chin J Appl Environ Biol, 2022, 28(4): 920-930. |
| 王媚, 刘天鹏, 何继红, 董孔军, 任瑞玉, 张磊, 杨天育. 糜子bZIP基因家族鉴定及幼苗期聚乙二醇6000处理下的表达特征. 应用与环境生物学报, 2022, 28(4): 920-930. | |
| [30] | Xin XX, Zheng XR, Wang HG, Chen L, Dipak KS, Wang RY, Qiao ZJ. Cloning and bioinformatics analysis of PmNAC1 in broomcorn millet. J Shanxi Agric Sci, 2023, 51(10):1162-1169. |
| 辛旭霞, 郑香然, 王海岗, 陈凌, Santra Dipak K, 王瑞云, 乔治军. 糜子PmNAC1的克隆及生物信息学分析. 山西农业科学, 2023, 51(10): 1162-1169. | |
| [31] | Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador- Vegas A, Salazar GA, Tate J, Bateman A. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res, 2016, 44(D1): D279-D285. |
| [32] |
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol, 2016, 33(7): 1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [33] | Ren RY, He JH, Dong KJ, Zhang L, Liu TP, Yang TY. Report on new-bred broomcorn millet cultivar Longmi 12. Gansu Agric Sci Technol, 2017, (3): 14-16. |
| 任瑞玉, 何继红, 董孔军, 张磊, 刘天鹏, 杨天育. 糜子新品种陇糜12号选育报告. 甘肃农业科技, 2017,(3): 14-16. | |
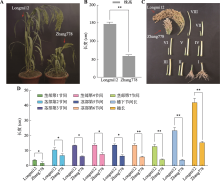
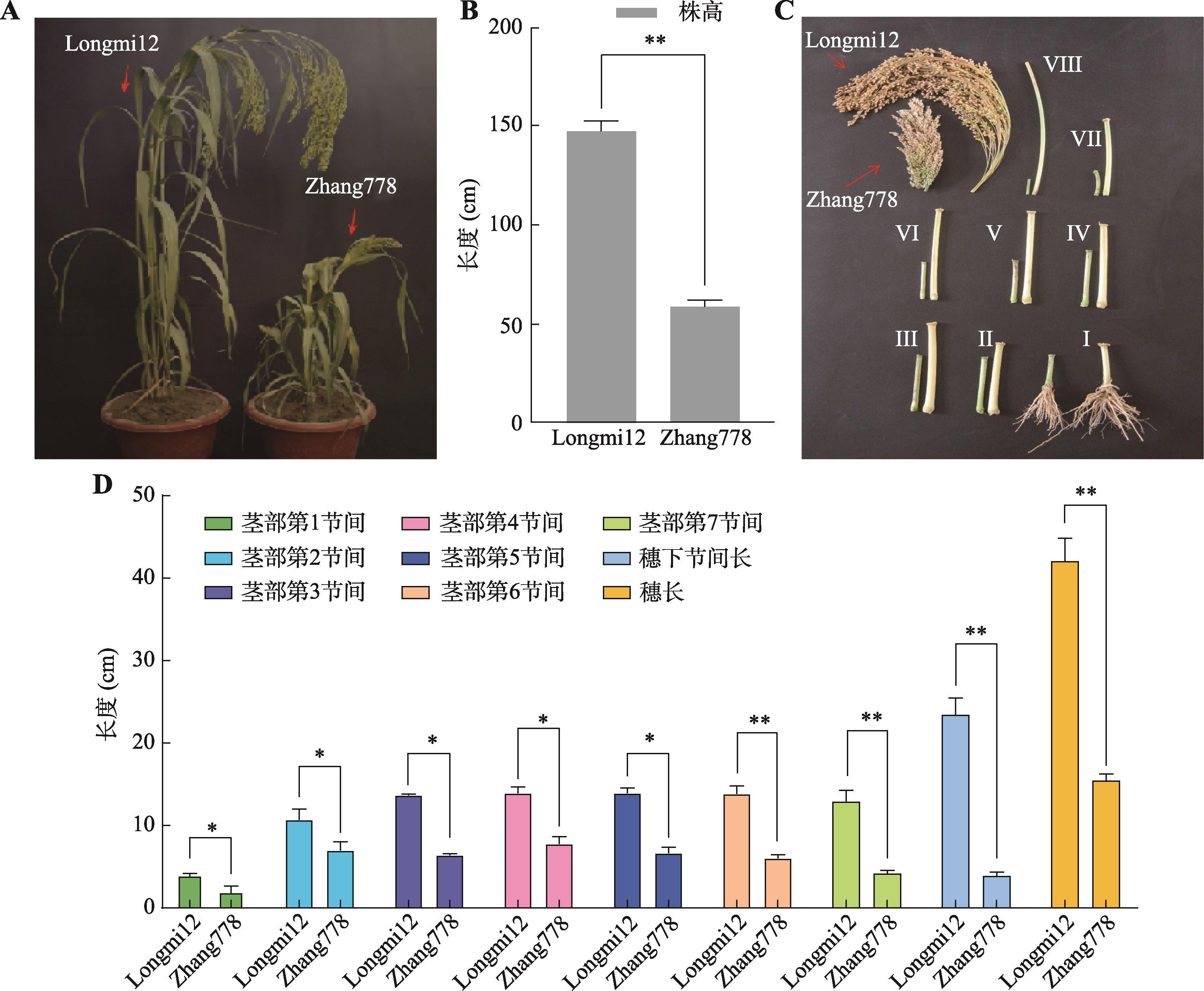
| [34] | Zhang B, Jia XP, Yang DZ, Zhao Y, Dai LF, Kou SJ, Zhang XM, Hou DY, Zhu XH. Investigation on agronomic characters of dwarf mutant in Panicum miliaceuml and analysis of its sensitivity to GA. Acta Agric Zhejiangensis, 2019, 31(5): 688-694. |
|
张博, 贾小平, 杨德智, 赵渊, 戴凌峰, 寇淑君, 张小梅, 侯典云, 朱学海. 糜子矮秆突变体778农艺性状调查及其对GA的敏感性分析. 浙江农业学报, 2019, 31(5): 688-694.
doi: 10.3969/j.issn.1004-1524.2019.05.02 |
|
| [35] |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods, 2001, 25: 402-408.
doi: 10.1006/meth.2001.1262 pmid: 11846609 |
| [36] |
Cao JF, Huang JQ, Liu X, Huang CC, Zheng ZS, Zhang XF, Shangguan XX, Wang LJ, Zhang YG, Wendel JF, Grover CE, Chen ZW. Genome-wide characterization of the GRF family and their roles in response to salt stress in Gossypium. BMC Genomics, 2020, 21(1): 1-16.
doi: 10.1186/s12864-019-6419-1 |
| [37] | Wang PJ, Zheng YC, Lin Y, Zhou Z, Yang JF, Ye NX. Genome-wide identification and expression analysis of GRF gene family in Camellia sinensis. Acta Bot Boreali- Occidentalia Sin, 2019, 39(3): 413-421. |
| 王鹏杰, 郑玉成林浥, 周珍, 杨江帆, 叶乃兴. 茶树GRF基因家族的全基因组鉴定及表达分析. 西北植物学报, 2019, 39(3): 413-421. | |
| [38] |
Zafar I, Rubab A, Aslam M, Ahmad SU, Liyaqat I, Malik A, Alam M, Wani TA, Khan AA. Genome-wide identification and analysis of GRF (growth-regulating factor) gene family in Camila sativa through in silico approaches. J King Saud Univ-Sci, 2022, 34(4): 102038.
doi: 10.1016/j.jksus.2022.102038 |
| [39] |
Wang FD, Qiu NW, Ding Q, Li JJ, Zhang YH, Li HY, Gao JW. Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genomics, 2014, 15(1): 1-12.
doi: 10.1186/1471-2164-15-1 |
| [1] | Yan Guo, Lele Yang, Huayu Qi. Transcriptome analysis of mouse male germline stem cells reveals characteristics of mature spermatogonial stem cells [J]. Hereditas(Beijing), 2022, 44(7): 591-608. |
| [2] | Sanzeng Zhao, Danyu Kong, Peiyong Xin, Jinfang Chu, Yinglang Wan, Hong-Qing Ling, Yi Liu. AtCPS V326M significantly affect the biosynthesis of gibberellins [J]. Hereditas(Beijing), 2022, 44(3): 245-252. |
| [3] | Hongbo Luo, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of a DNA methylation-driven transcriptional signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2020, 42(8): 775-787. |
| [4] | Tianpei Shi,Li Zhang. Application of whole transcriptomics in animal husbandry [J]. Hereditas(Beijing), 2019, 41(3): 193-205. |
| [5] | Gaohua Zhang, Shutao Yu, He Wang, Xuda Wang. Transcriptome profiling of high oleic peanut under low temperatureduring germination [J]. Hereditas(Beijing), 2019, 41(11): 1050-1059. |
| [6] | Lan Ren,Rudan Xiao,Qian Zhang,Xiaomin Lou,Zhaojun Zhang,Xiangdong Fang. Synergistic regulation of the erythroid differentiation of K562 cells by KLF1 and KLF9 [J]. Hereditas(Beijing), 2018, 40(11): 998-1006. |
| [7] | Yajun Liu,Feng Zhang,Hongde Liu,Xiao Sun. The application of next-generation sequencing techniques in studying transcriptional regulation in embryonic stem cells [J]. Hereditas(Beijing), 2017, 39(8): 717-725. |
| [8] | Kai Wei,Lei Ma. Concept development of housekeeping genes in the high-throughput sequencing era [J]. Hereditas(Beijing), 2017, 39(2): 127-134. |
| [9] | Guangqi Li, Congjiao Sun, Guiqin Wu, Fengying Shi, Aiqiao Liu, Hao Sun, Ning Yang. Transcriptome sequencing identifies potential regulatory genes involved in chicken eggshell brownness [J]. Hereditas(Beijing), 2017, 39(11): 1102-1111. |
| [10] | Yongming Liu, Ling Zhang, Tao Qiu, Zhuofan Zhao, Moju Cao. Research progress on mechanisms of male sterility in plants based on high-throughput RNA sequencing [J]. Hereditas(Beijing), 2016, 38(8): 677-687. |
| [11] | Xiao Zhang, Guifang Jia. RNA epigenetic modification: N6-methyladenosine [J]. HEREDITAS(Beijing), 2016, 38(4): 275-288. |
| [12] | Shuaiqi Zhu, Yifu Gong, Yuqing Hang, Hao Liu, Heyu Wang. Transcriptome analysis of Dunaliella viridis [J]. HEREDITAS(Beijing), 2015, 37(8): 828-836. |
| [13] | Dong Wang, Yongjun Li, Nan Ding, Junyun Wang, Qiong Yang, Yaran Yang, Yanming Li, Xiangdong Fang, Hua Zhao. Molecular networks and mechanisms of epithelial-mesenchymal transition regulated by miRNAs in the malignant melanoma cell line [J]. HEREDITAS(Beijing), 2015, 37(7): 673-682. |
| [14] | Wei Jiang, Yixing Fan, Xian Qiao, Yanjun Zhang, Zhihong Liu, Yanhong Zhao, Ruijun Wang, Zhixin Wang, Wenguang Zhang, Rui Su, Jinquan Li. The transcriptome research progresses of skin hair follicle development [J]. HEREDITAS(Beijing), 2015, 37(6): 528-534. |
| [15] | Yongming Liu, Ling Zhang, Jianyu Zhou, Moju Cao. Research progress of the bHLH transcription factors involved in genic male sterility in plants [J]. HEREDITAS(Beijing), 2015, 37(12): 1194-1203. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||