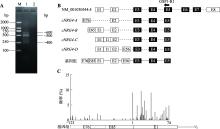
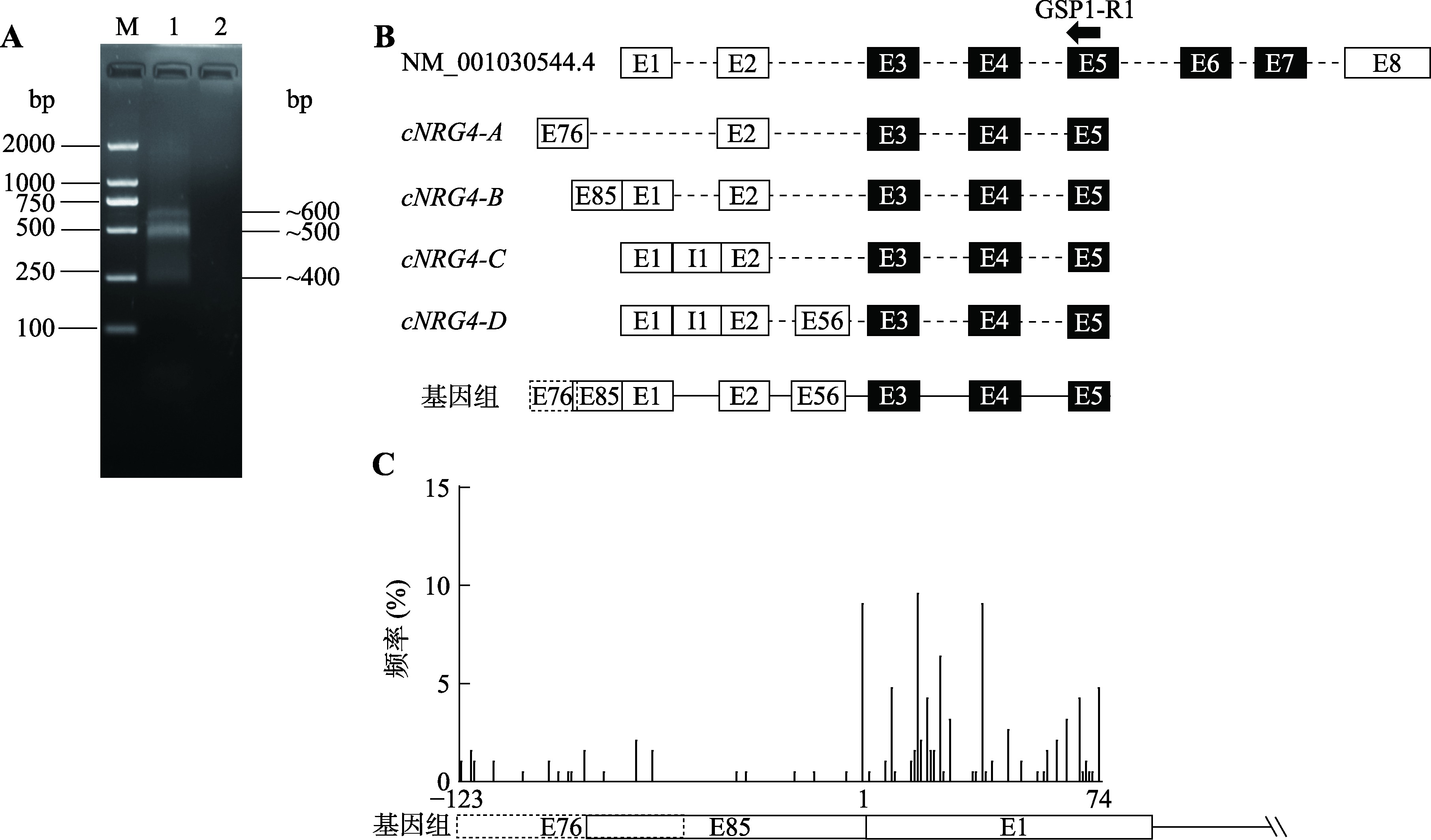
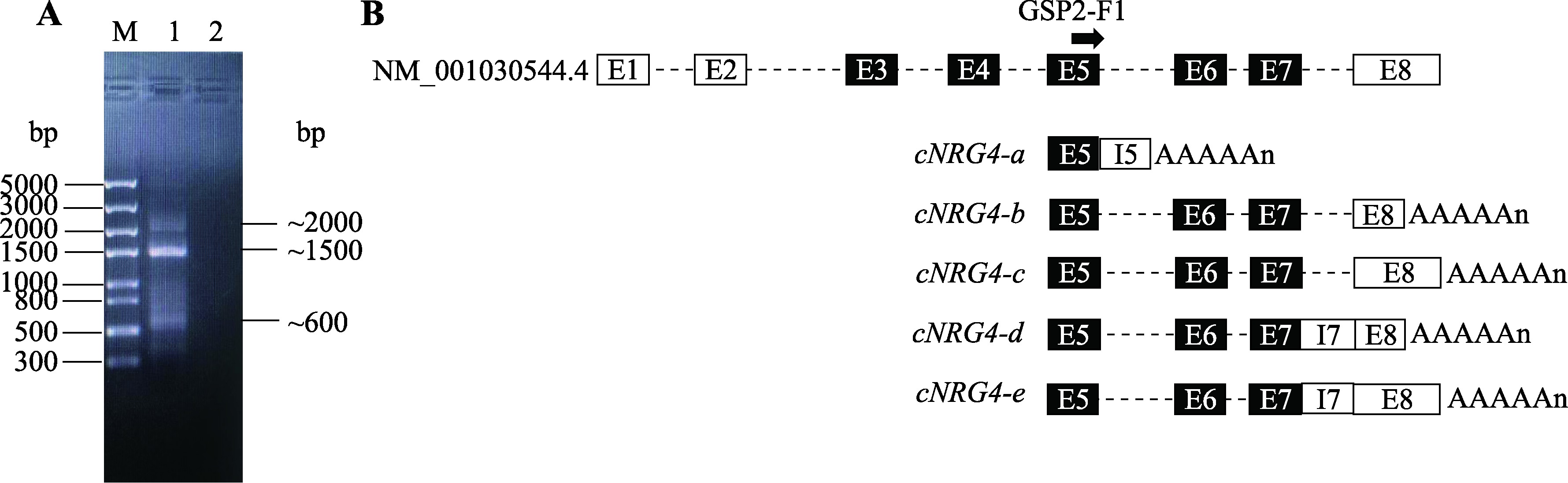
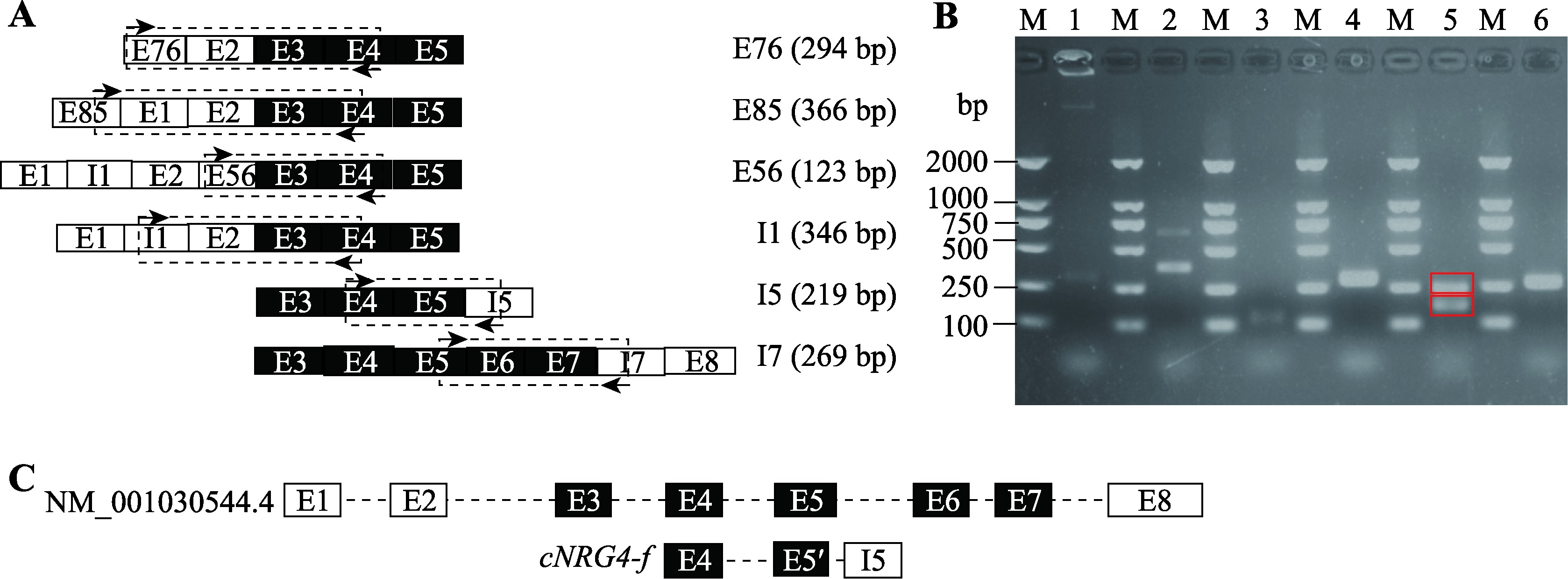
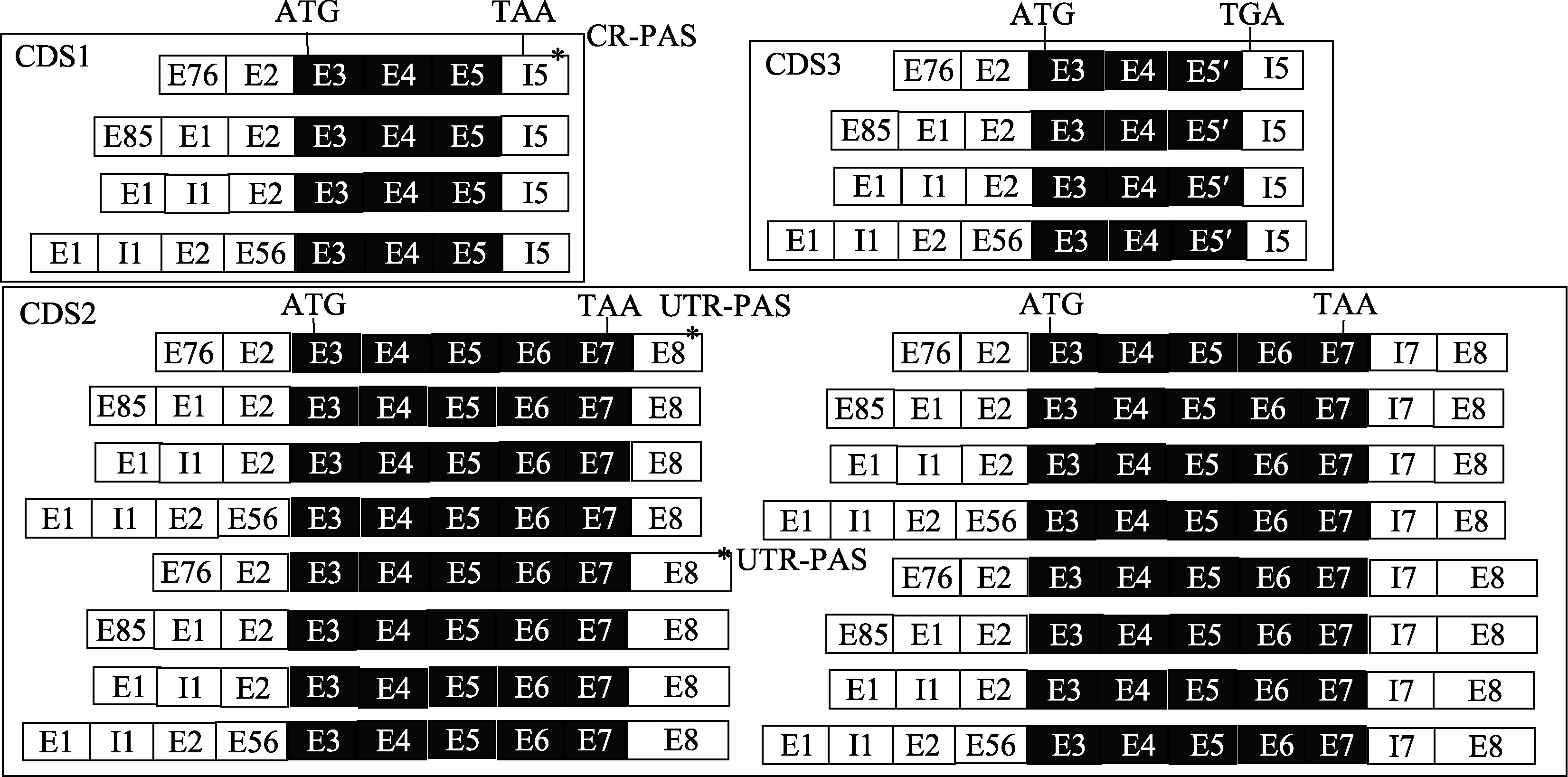
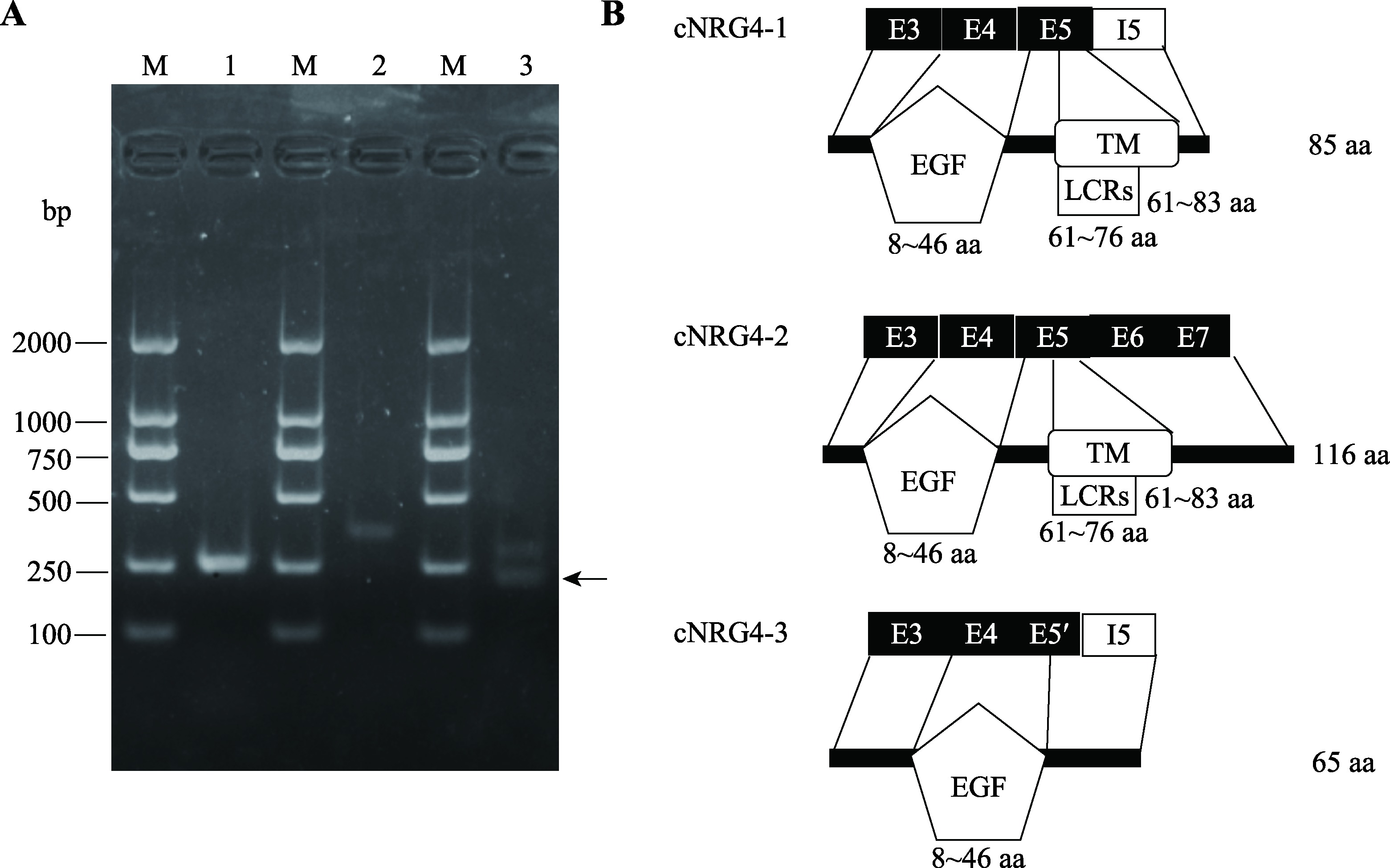
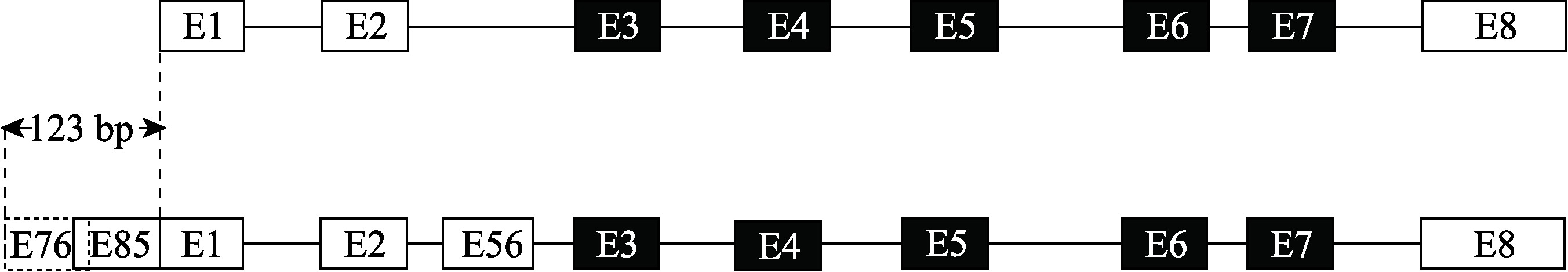
Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (5): 447-458.doi: 10.16288/j.yczz.23-001
• Research Article • Previous Articles
Characterization of the genomic and transcriptional structure of chicken NRG4 gene
Zhihui Gao1,2,3( ), Jiaxin Huang1,2,3, Haoyu Luo1,2,3, Haidong Xu1,2,3, Ming Lou1,2,3, Bolin Ning1,2,3, Xiaoxu Xing1,2,3, Fang Mu1,2,3, Hui Li1,2,3, Ning Wang1,2,3(
), Jiaxin Huang1,2,3, Haoyu Luo1,2,3, Haidong Xu1,2,3, Ming Lou1,2,3, Bolin Ning1,2,3, Xiaoxu Xing1,2,3, Fang Mu1,2,3, Hui Li1,2,3, Ning Wang1,2,3( )
)
- 1. Key Laboratory of Chicken Genetics and Breeding, Ministry of Agriculture and Rural Affairs, Harbin 150030, China
2. Key Laboratory of Animal Genetics, Breeding and Reproduction, Education Department of Heilongjiang Province, Harbin 150030, China
3. College of Animal Science and Technology, Northeast Agricultural University, Harbin 150030, China
-
Received:2023-01-03Revised:2023-04-04Online:2023-05-20Published:2023-04-19 -
Contact:Wang Ning E-mail:gaozhihui@neau.edu.cn;wangning@neau.edu.cn -
Supported by:National Natural Science Foundation of China(31872346);China Agriculture Research System(CARS-41)
Cite this article
Zhihui Gao, Jiaxin Huang, Haoyu Luo, Haidong Xu, Ming Lou, Bolin Ning, Xiaoxu Xing, Fang Mu, Hui Li, Ning Wang. Characterization of the genomic and transcriptional structure of chicken NRG4 gene[J]. Hereditas(Beijing), 2023, 45(5): 447-458.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Primers used in this study"
| 引物类型 | 引物名称 | 引物序列(5?→3?) |
|---|---|---|
| RACE | GSP1-R1 | AGAAGGAGAAGTGAAGCCAAGAGAACTG |
| GSP2-F1 | GTTTGCAGTTCTCTTGGCTTCACTTCTC | |
| RT-PCR | E76-F/E4-R2 | GCCGAGCCAATGGGCGACG GATCCATAACTGGTGCCACAGAGT |
| E85-F/E4-R1 | GGTGAGGCGCGAGAG GACAAAAAGATCCATAACTGGTGC | |
| E56-F/E4-R2 | CTCCTTCACAGGCACAGTTCAG GATCCATAACTGGTGCCACAGAGT | |
| I1-F/E4-R1 | CTCTGTTAACGTTCCGCCTG GACAAAAAGATCCATAACTGGTGC | |
| E4-F /I5-R | TTCCTACTGTACCCAGTCCATTC CTACTTGATTAAATAGCTTCAGCAC | |
| E5-F /I7-R | CTTCTCGGAGTTCTTTTAATTGGA TCTCTAAGATAAAAGTAGGCCTC | |
| CDS1-F/R | ATGCGAACAGATCATGAAG TTACCTGCAAAGGAAGTAG | |
| CDS2-F/R | ATGCGAACAGATCATGAAG TTACTCCGTTATTATTTTGTTGCAG | |
| CDS3-F/R | ATGCGAACAGATCATGAAG TAGTACATGATTACCAGAAGCAA |
| [1] |
Yang F, Li XN. Research progress of neuregulin 4 biological function. Acta Physiol Sin, 2017, 69(3): 351-356.
pmid: 28638929 |
|
杨帆, 李晓南. 神经调节蛋白4生物学功能的研究进展. 生理学报, 2017, 69(3): 351-356.
pmid: 28638929 |
|
| [2] |
Ledonne A, Mercuri NB. On the modulatory roles of neuregulins/ErbB signaling on synaptic plasticity. Int J Mol Sci, 2019, 21(1): 275.
doi: 10.3390/ijms21010275 |
| [3] |
Dai YN, Zhu JZ, Fang ZY, Zhao DJ, Wan XY, Zhu HT, Yu CH, Li YM. A case-control study: association between serum neuregulin 4 level and non-alcoholic fatty liver disease. Metabolism, 2015, 64(12): 1667-1673.
doi: 10.1016/j.metabol.2015.08.013 |
| [4] |
Wang GX, Zhao XY, Meng ZX, Kern M, Dietrich A, Chen ZM, Cozacov Z, Zhou DQ, Okunade AL, Su X, Li SM, Blüher M, Lin JD. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med, 2014, 20(12): 1436-1443.
doi: 10.1038/nm.3713 |
| [5] |
Chen LL, Peng MM, Zhang JY, Hu X, Min J, Huang QL, Wan LM. Elevated circulating Neuregulin4 level in patients with diabetes. Diabetes Metab Res Rev, 2017, 33(4): e2870.
doi: 10.1002/dmrr.2870 |
| [6] |
Steinthorsdottir V, Stefansson H, Ghosh S, Birgisdottir B, Bjornsdottir S, Fasquel AC, Olafsson O, Stefansson K, Gulcher JR.Multiple novel transcription initiation sites for NRG1. Gene, 2004, 342(1): 97-105.
pmid: 15527969 |
| [7] |
Rimer M, Prieto AL, Weber JL, Colasante C, Ponomareva O, Fromm L, Schwab MH, Lai C, Burden SJ.Neuregulin-2 is synthesized by motor neurons and terminal Schwann cells and activates acetylcholine receptor transcription in muscle cells expressing ErbB4. Mol Cell Neurosci, 2004, 26(2): 271-281.
pmid: 15207852 |
| [8] |
Hayes NVL, Gullick WJ. The neuregulin family of genes and their multiple splice variants in breast cancer. J Mammary Gland Biol Neoplasia, 2008, 13(2): 205-214.
doi: 10.1007/s10911-008-9078-4 |
| [9] |
Carteron C, Ferrer-Montiel A, Cabedo H. Characterization of a neural-specific splicing form of the human neuregulin 3 gene involved in oligodendrocyte survival. J Cell Sci, 2006, 119(Pt 5): 898-909.
doi: 10.1242/jcs.02799 pmid: 16478787 |
| [10] |
Hayes NVL, Newsam RJ, Baines AJ, Gullick WJ. Characterization of the cell membrane-associated products of the neuregulin 4 gene. Oncogene, 2008, 27(5): 715-720.
pmid: 17684490 |
| [11] |
Hayes NVL, Blackburn E, Smart LV, Boyle MM, Russell GA, Frost TM, Morgan BJT, Baines AJ, Gullick WJ. Identification and characterization of novel spliced variants of neuregulin 4 in prostate cancer. Clin Cancer Res, 2007, 13(11): 3147-3155.
doi: 10.1158/1078-0432.CCR-06-2237 pmid: 17545517 |
| [12] | Guo YQ, Wang WJ, Gao ZH, Mu F, Xu HD, Li H, Wang N. Cloning, expression and promoter analysis of adipokine NRG4 gene in chicken. Chin J Agric Biotechol, 2021, 29(11): 2129-2138. |
| 郭亚琦, 王伟佳, 高智慧, 牟芳, 徐海冬, 李辉, 王宁. 鸡脂肪细胞因子NRG4基因的克隆、表达及启动子分析. 农业生物技术学报, 2021, 29(11): 2129-2138. | |
| [13] |
Pfeifer A. NRG4: an endocrine link between brown adipose tissue and liver. Cell Metab, 2015, 21(1): 13-14.
doi: 10.1016/j.cmet.2014.12.008 pmid: 25565202 |
| [14] | Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol, 2018, 19(10): 621-637. |
| [15] |
Forutan M, Ross E, Chamberlain AJ, Nguyen L, Mason B, Moore S, Garner JB, Xiang RD, Hayes BJ. Evolution of tissue and developmental specificity of transcription start sites in Bos taurus indicus. Commun Biol, 2021, 4(1): 829.
doi: 10.1038/s42003-021-02340-6 pmid: 34211114 |
| [16] |
Mejía-Guerra MK, Li W, Galeano NF, Vidal M, Gray J, Doseff AI, Grotewold E. Core promoter plasticity between maize tissues and genotypes contrasts with predominance of sharp transcription initiation sites. Plant Cell, 2015, 27(12): 3309-3320.
doi: 10.1105/tpc.15.00630 |
| [17] |
Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engström PG, Frith MC, Forrest ARR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet, 2006, 38(6): 626-635.
doi: 10.1038/ng1789 pmid: 16645617 |
| [18] |
Thieffry A, López-Márquez D, Bornholdt J, Malekroudi MG, Bressendorff S, Barghetti A, Sandelin A, Brodersen P. PAMP-triggered genetic reprogramming involves widespread alternative transcription initiation and an immediate transcription factor wave. Plant Cell, 2022, 34(7): 2615-2637.
doi: 10.1093/plcell/koac108 |
| [19] |
Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res, 2014, 24(11): 1774-1786.
doi: 10.1101/gr.177790.114 pmid: 25258385 |
| [20] |
Hammarskjöld ML. Regulation of retroviral RNA export. Semin Cell Dev Biol, 1997, 8(1): 83-90.
pmid: 15001110 |
| [21] |
Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in arabidopsis. Plant J, 2004, 39(6): 877-885.
doi: 10.1111/j.1365-313X.2004.02172.x pmid: 15341630 |
| [22] | Rekosh D, Hammarskjold ML. Intron retention in viruses and cellular genes: detention, border controls and passports. Wiley Interdiscip Rev RNA, 2018, 9(3): e1470. |
| [23] |
Marquez Y, Höpfler M, Ayatollahi Z, Barta A, Kalyna M. Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res, 2015, 25(7): 995-1007.
doi: 10.1101/gr.186585.114 pmid: 25934563 |
| [24] |
Tahmasebi S, Jafarnejad SM, Tam IS, Gonatopoulos- Pournatzis T, Matta-Camacho E, Tsukumo Y, Yanagiya A, Li WC, Atlasi Y, Caron M, Braunschweig U, Pearl D, Khoutorsky A, Gkogkas CG, Nadon R, Bourque G, Yang XJ, Tian B, Stunnenberg HG, Yamanaka Y, Blencowe BJ, Giguère V, Sonenberg N.Control of embryonic stem cell self-renewal and differentiation via coordinated alternative splicing and translation of YY2. Proc Natl Acad Sci USA, 2016, 113(44): 12360-12367.
pmid: 27791185 |
| [25] |
Weatheritt RJ, Sterne-Weiler T, Blencowe BJ. The ribosome-engaged landscape of alternative splicing. Nat Struct Mol Biol, 2016, 23(12): 1117-1123.
doi: 10.1038/nsmb.3317 pmid: 27820807 |
| [26] |
Sun SY, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post- transcriptional and translational control. Nat Struct Mol Biol, 2010, 17(3): 306-312.
doi: 10.1038/nsmb.1750 |
| [27] |
Thiele A, Nagamine Y, Hauschildt S, Clevers H. AU-rich elements and alternative splicing in the beta-catenin 3'UTR can influence the human beta-catenin mRNA stability. Exp Cell Res, 2006, 312(12): 2367-2378.
pmid: 16696969 |
| [28] |
Nourse J, Spada S, Danckwardt S. Emerging roles of RNA 3'-end cleavage and polyadenylation in pathogenesis, diagnosis and therapy of human disorders. Biomolecules, 2020, 10(6): 915.
doi: 10.3390/biom10060915 |
| [29] |
Chen W, Jia Q, Song YF, Fu HH, Wei G, Ni T. Alternative polyadenylation: methods, findings, and impacts. Genomics Proteomics Bioinformatics, 2017, 15(5): 287-300.
doi: 10.1016/j.gpb.2017.06.001 |
| [30] |
Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA, 2007, 13(5): 625-642.
pmid: 17449729 |
| [31] |
Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol, 2017, 18(1): 18-30.
doi: 10.1038/nrm.2016.116 |
| [32] |
Lau JS, Yip CW, Law KM, Leung FC. Cloning and characterization of chicken growth hormone binding protein (cGHBP). Domest Anim Endocrinol, 2007, 33(1): 107-121.
doi: 10.1016/j.domaniend.2006.04.012 |
| [33] |
Ning BL, Huang JX, Xu HD, Lou YQ, Wang WS, Mu F, Yan XH, Li H, Wang N. Genomic organization, intragenic tandem duplication, and expression analysis of chicken TGFBR2 gene. Poult Sci, 2022, 101(12): 102169.
doi: 10.1016/j.psj.2022.102169 |
| [34] |
Wu QS, Wright M, Gogol MM, Bradford WD, Zhang N, Bazzini AA. Translation of small downstream ORFs enhances translation of canonical main open reading frames. EMBO J, 2020, 39(17): e104763.
doi: 10.15252/embj.2020104763 |
| [35] |
Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, Vejnar CE, Lee MT, Rajewsky N, Walther TC, Giraldez AJ. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J, 2014, 33(9): 981-993.
doi: 10.1002/embj.201488411 pmid: 24705786 |
| [36] |
Dodbele S, Wilusz JE. Ending on a high note: downstream ORFs enhance mRNA translational output. EMBO J, 2020, 39(17): e105959.
doi: 10.15252/embj.2020105959 |
| [1] | Xian Zou, Yanhua He, Jingyi He, Yan Wang, Dingming Shu, Chenglong Luo. Optimization of transfection conditions of chicken primordial germ cells [J]. Hereditas(Beijing), 2021, 43(3): 280-288. |
| [2] | Bingyuan Wang, Yulian Mu, Kui Li, Zhiguo Liu. Research progress of stem cells in agricultural animals [J]. Hereditas(Beijing), 2020, 42(11): 1073-1080. |
| [3] | Qiying Leng, Jiahui Zheng, Haidong Xu, Patricia Adu-Asiamah, Ying Zhang, Bingwang Du, Li Zhang. Cloning and expression analysis of chicken circular transcript of insulin degrading enzyme gene [J]. Hereditas(Beijing), 2019, 41(12): 1129-1137. |
| [4] | Jiahui Chen, Xueyi Ren, min Li, Shiyi Lu, Tian Cheng, Liangtian Tan, Shaodong Liang, Danlin He, Qingbin Luo, Qinghua Nie, Xiquan Zhang, Wen Luo. The cell cycle pathway regulates chicken abdominal fat deposition as revealed by transcriptome sequencing [J]. Hereditas(Beijing), 2019, 41(10): 962-973. |
| [5] | Yankai Chu,Yanfei Jin,Tianyu Xing,Guangwei Ma,Tingting Cui,Xiaohong Yan,Hui Li,Ning Wang. Post-transcriptional regulation of chicken PPARγ transcript variant 3 by upstream open reading frame [J]. Hereditas(Beijing), 2018, 40(8): 657-667. |
| [6] | Xiaofei Zhang,He Song,Jing Liu,Wenjian Zhang,Xiaohong Yan,Hui Li,Ning Wang. Identification and analysis of ZFPM2 as a target gene of miR-17-92 cluster in chicken [J]. Hereditas(Beijing), 2017, 39(4): 333-345. |
| [7] | Guangqi Li, Congjiao Sun, Guiqin Wu, Fengying Shi, Aiqiao Liu, Hao Sun, Ning Yang. Transcriptome sequencing identifies potential regulatory genes involved in chicken eggshell brownness [J]. Hereditas(Beijing), 2017, 39(11): 1102-1111. |
| [8] | Min Cheng, Wenjian Zhang, Tianyu Xing, Xiaohong Yan, Yumao Li, Hui Li, Ning Wang. Functional analysis of the upstream regulatory region of chicken miR-17-92 cluster [J]. Hereditas(Beijing), 2016, 38(8): 724-735. |
| [9] | Tao Zhang, Wenhao Wang, Genxi Zhang, Jinyu Wang, Qian Xue, Yuping Gu. A genome-wide association study on body weight traits of Jinghai yellow chicken [J]. HEREDITAS(Beijing), 2015, 37(8): 811-820. |
| [10] | Junping Ma, Xi Yang, Na Lv, Fei Liu, Yan Chen, Baoli Zhu. Re-sequencing and assembly of chicken T cell receptor gamma locus [J]. HEREDITAS(Beijing), 2015, 37(6): 568-574. |
| [11] | Xiaoyan Song, Dexiang Zhang, Wenwu Zhang, Congliang Ji, Xiquan Zhang, Qingbin Luo. Gene expression profiling of three tissues of chicken after heat stress treatment by microarray technique [J]. HEREDITAS(Beijing), 2014, 36(8): 800-808. |
| [12] | Jinlong Li, Shaoqing Tang, Zhiyuan Zou, Haichao Wang, Qing Xu. Establishment of fluorescence labeling and capillary electrophoresis in MSAP for Beijing You chicken [J]. HEREDITAS(Beijing), 2014, 36(5): 495-502. |
| [13] | MU Ren BIAN Yan-Chao PU Ya-Bin LI Xiang-Chen WANG Feng-Long GUAN Wei-Jun. Isolation and biological characterization of mesenchymal stem cells from Beijing fatty chicken fetal liver [J]. HEREDITAS, 2013, 35(3): 365-372. |
| [14] | WANG Li, NA Wei, WANG Yu-Xiang, WANG Yan-Bo, WANG Ning, WANG Qi-Gui, LI Yu-Mao, LI Hui. Characterization of chicken PPARγ expression and its impact on adipocyte proliferation and differentiation [J]. HEREDITAS, 2012, 34(4): 454-464. |
| [15] | . Association of single nucleotide polymorphism of RB1 gene with body weight traits in chicken [J]. HEREDITAS, 2012, 34(10): 1320-1327. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||