遗传 ›› 2022, Vol. 44 ›› Issue (8): 655-671.doi: 10.16288/j.yczz.22-158
收稿日期:2022-05-15
修回日期:2022-06-28
出版日期:2022-08-20
发布日期:2022-07-12
通讯作者:
蒋婧
E-mail:tian.xie@sibcb.ac.cn;jiangjing@sibcb.ac.cn
作者简介:谢甜,硕士,工程师,研究方向:基因组标签计划与基因编辑。E-mail: 基金资助:
Tian Xie( ), Mei Wang, Ruiyu Gao, Yanni Miao, Yiming Zhang, Jing Jiang(
), Mei Wang, Ruiyu Gao, Yanni Miao, Yiming Zhang, Jing Jiang( )
)
Received:2022-05-15
Revised:2022-06-28
Online:2022-08-20
Published:2022-07-12
Contact:
Jiang Jing
E-mail:tian.xie@sibcb.ac.cn;jiangjing@sibcb.ac.cn
Supported by:摘要:
位点特异性重组系统由重组酶和特异性识别位点两部分组成,是一种强大的基因操作工具,被广泛运用于生命科学研究。已开发的诱导型重组系统以时空方式精准调控细胞和动物的基因表达,被用于基因功能研究、细胞谱系示踪和疾病治疗等领域。根据诱导重组酶时空表达方式的不同,诱导型重组系统可分为化学诱导和光控诱导两种方式。光控诱导重组系统是利用光作为诱导剂,根据光控方式和对象的不同,可进一步分为光笼和光遗传学两类。光笼诱导重组系统是利用光敏基团来控制化学诱导剂或重组酶,光诱导前它们的活性被光敏基团抑制;在特定光照射后,它们的活性被恢复,进而实现光控诱导基因重组。光遗传学诱导重组系统是通过光遗传学开关介导分割型重组酶的重新激活来诱导基因重组。其中光遗传学开关由一系列基因编码的光敏蛋白组成,包括隐花色素、VIVID蛋白、光敏色素等。这些类型丰富的光控诱导重组系统为从高时空分辨率的维度解析基因的表达和功能提供了更多的工具,以满足日益复杂的生命科学研究需求。本文主要对不同类型光控诱导重组系统的开发原理及应用进行综述,比较其优缺点,最后对未来开发更多光控重组系统进行展望, 旨在为系统优化升级提供理论基础和指导。
谢甜, 王梅, 高瑞钰, 苗艳尼, 张燚铭, 蒋婧. 光控诱导重组系统的开发与应用[J]. 遗传, 2022, 44(8): 655-671.
Tian Xie, Mei Wang, Ruiyu Gao, Yanni Miao, Yiming Zhang, Jing Jiang. Development and application of light-controlled inducible recombination systems[J]. Hereditas(Beijing), 2022, 44(8): 655-671.
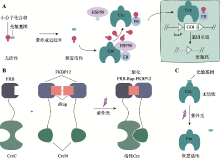
图1
基于光笼分子的光控诱导重组系统 A:光笼化学诱导剂。在光控诱导CreER/loxP系统中,经光笼修饰的小分子化合物活性被抑制。在特定波段的光照下,光控释放的小分子化合物恢复活性,使得CreER融合蛋白与HSP90发生解离。CreER被转运进入细胞核后可识别loxP序列,诱导两个loxP序列之间的目标基因(gene of interest,GOI)发生重组。B:光裂解雷帕霉素二聚体dRap。紫外光照射后,dRap裂解释放天然雷帕霉素Rap,从而诱导FRB-CreC和FKBP12-CreN二聚化,使得分割型Cre重组酶重构恢复催化活性。C:光笼重组酶。紫外光照射后,光笼Cre重组酶恢复活性。"
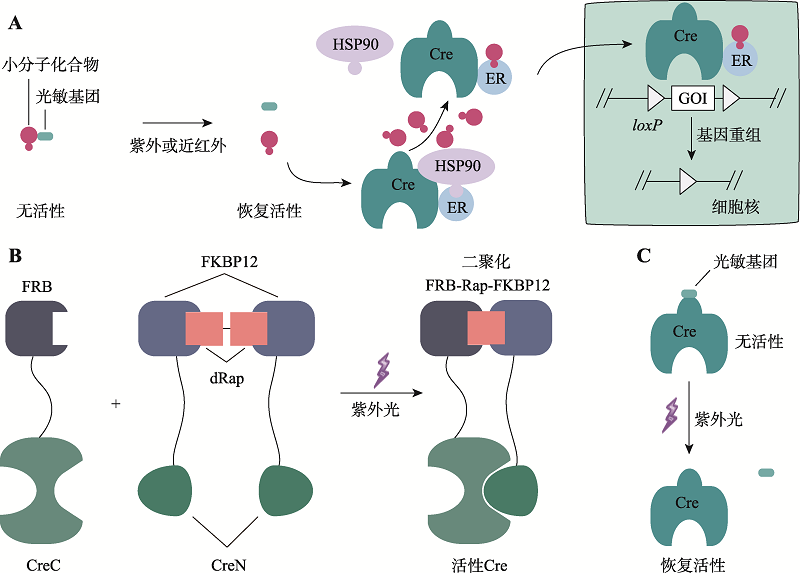
表1
光笼诱导重组系统的比较"
| 光笼修饰底物 | 光敏基团 | 诱导光类型 | 优点 | 缺点 | 参考文献 |
|---|---|---|---|---|---|
| 4-羟基他莫昔芬氮丙啶 | 4,5-二甲氧基-2-硝基苯甲醇 | 紫外光 365 nm | 可时空调控 | 存在背景泄露,重组效率较低,缺乏在体实验 | [ |
| 4-羟基环芬 | 4,5-二甲氧基-2-硝基苯甲醇 | 紫外光 365 nm | 光笼易合成、易溶于水、产量高,系统具有高时空分辨率 | 紫外光组织穿透能力较差 | [ |
| 他莫昔芬 | 邻硝基苄基 | 紫外光 365 nm | 光笼易合成、易溶于水,短时间紫外暴露即可释放他莫昔芬 | 应用仅限于部分细胞,存在背景泄露,重组效率较低,缺乏在体实验 | [ |
| 4-羟基他莫昔芬 | 邻硝基苄基 | 紫外光 365 nm | 光笼活性高,系统敏感、具有高时空分辨率 | 存在背景泄露,缺乏在体实验 | [ |
| 雷帕霉素 | N,N′-二琥珀酰亚胺基碳酸酯 | 紫外光 365 nm | 对光敏感,设置简易,精准时空调控 | 雷帕霉素对生物体内蛋白活性有影响,缺乏在体实验 | [ |
| 4-羟基环芬 | 花菁 | 近红外光 690 nm | 光毒性低,组织穿透性强,可以特异性传递小分子化合物 | 缺乏在体实验 | [ |
| Cre重组酶 | 邻硝基苄基 | 紫外光 365 nm | 由基因编码,无需额外小分子化合物诱导,严格响应光调控 | 光笼重组酶蛋白获取困难,缺乏在体实验 | [ |
表2
光遗传学诱导重组系统的比较"
| 光遗传学诱 导重组系统 | 诱导光类型 | 光遗传学元件 | 重组酶模块 | 优点 | 缺点 | 参考文献 |
|---|---|---|---|---|---|---|
| PA-Cre 1.0 | 蓝光 450 nm | CRY2 (aa:1~612) | CreN (aa:19~104) | 亚秒时间和亚细胞空间分辨率,可逆性 | 组织穿透力不足,背景泄露高 | [ |
| CIBN (aa:1~170) | CreC (aa:106~343) | |||||
| PA-Cre 1.5 | 蓝光 461 nm | CRY2(L348F) | CreN (aa:19~104) | 背景泄露较低,可逆性 | 组织穿透力不足 | [ |
| CIBN (aa:1~170) | CreC (aa:106~343) | |||||
| 改良型 PA-Cre 2.0 | 蓝光 461 nm | ER-CRY2(L348F) | CreN (aa:19~104) | 背景泄露较低,光敏感,诱导活性高,可逆性 | 需调控细胞核内与核外蛋白表达浓度,以获得低背景泄露 | [ |
| NLS-CIB1 (aa:1~335) | CreC (aa:106~343) | |||||
| Li-rtTA | 蓝光 470 nm | CIBN-rTetR-CIBN | TRE-Cre | 蓝光和强力霉素双重诱导,可逆性,时空特异性 | 小鼠繁殖复杂,耗时 | [ |
| CRY2PHR-VP16 | ||||||
| Magnets- PA-Cre | 蓝光 470±20 nm | nMag | CreN (aa:19~59) | 可逆性,重构的Cre可识别其他变体位点 | 背景泄露高,解聚慢 | [ |
| pMag | CreC (aa:60~343) | |||||
| Magnets- PA-Cre 3.0 | 蓝光 470±20 nm | nMag | CreN (aa:19~59) | 暗泄漏低,重组效率高,可逆性 | 组织穿透力不足 | [ |
| pMag | CreC (aa:60~343) | |||||
| TamPA- Cre | 蓝光 472±29 nm | nMag | ER-CreN (aa:2~59) | 蓝光和他莫昔芬双重诱导,光敏感,暗泄漏低,重组效率高,可逆性 | 他莫昔芬诱导后的核易位需要时间 | [ |
| NLS-pMag | CreC (aa:60~343) | |||||
| TRE-PA- Cre | 蓝光 470±20 nm | nMag | TRE-CreN (aa:19~59) | 蓝光和强力霉素双重诱导,可逆性 | 组织穿透力不足,重组效果待深入探究 | [ |
| pMag | CreC (aa:60~343) | |||||
| CreLite | 红光/远红光 640/750 nm | PhyB (aa:1~161) | CreC (aa:60~343) | 光敏感,光毒性低,组织穿透力强,可逆性 | 需要外源引入藻蓝胆素,重组效果待深入探究 | [ |
| PIF6 (aa:1~100) | CreN (aa:19~59) | |||||
| FISC | 远红光 730 nm | BphS | DocS-CreC (aa:60~343) | 无需辅助因子,组织穿透力强,光毒性低,重组效率高,背景泄露低 | 系统复杂,需要开发模块小装载能力大的载体,以确保体内有效递送 | [ |
| Coh2-CreN (aa:1~59) | ||||||
| PA-Flp | 蓝光 470 nm | nMagH | FlpN27 | 光敏感,背景泄露低,可与Cre重组酶交叉使用标记细胞亚群 | 组织穿透力不足 | [ |
| pMagH | FlpC28 | |||||
| PA-Dre | 蓝光 470 nm | nMag | DreN246 | 光敏感,背景泄露低,重构的Dre可识别rox及其变体,可与Cre重组酶交叉使用标记特定细胞 | 组织穿透力不足 | [ |
| pMag | DreC247 |
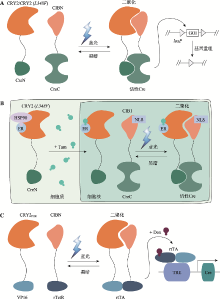
图2
基于隐花色素的光控诱导重组系统 A:CRY2或者CRY2(L348F)融合CreN,CIBN融合CreC构建的PA-Cre 1.0以及优化的PA-Cre 1.5系统。在黑暗条件下,Cre被分成两个片段对loxP位点没有催化活性。在蓝光照射下,CRY2或者CRY2(L348F)与CIBN发生二聚化介导CreN和CreC互补重构,使得Cre迅速恢复催化活性,可识别两个loxP位点发生DNA重组。B:光与他莫昔芬Tam双重调控的改良型PA-Cre 2.0系统,包含融合了ER的CRY2 (L348F)-CreN和NLS-CIB1-CreC,通过Tam控制核转运和光介导组装分割片段,实现对Cre重组酶活性的双重控制,提供更为复杂的DNA重组调控。C:Li-rtTA系统。rtTA的两个功能域即DNA结合域rTetR和转录激活结构域VP16分别与CIBN和CRY2PHR相融合。蓝光刺激CRY2PHR和CIBN的二聚化,促使rTetR和VP16组合发挥完整的rtTA功能。在强力霉素Dox存在的情况下,二聚化的融合蛋白激活Tet-on系统,驱动Cre的表达。NLS:核定位信号。"
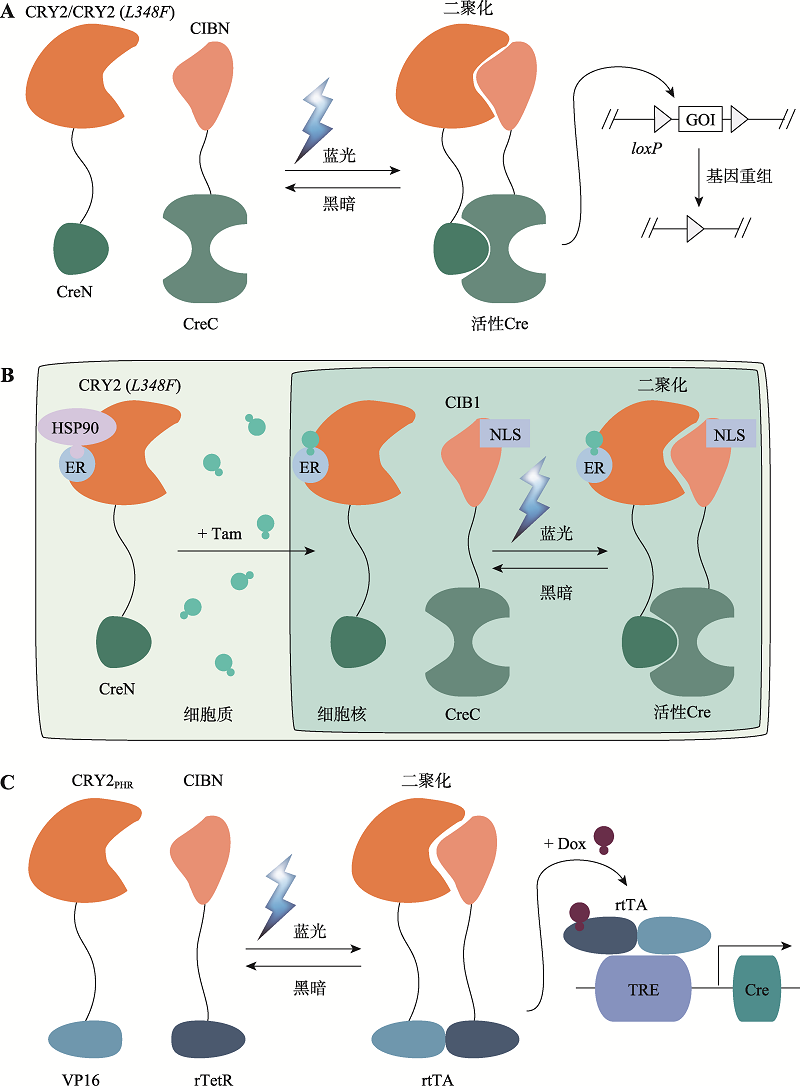
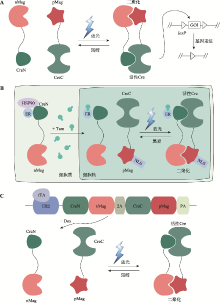
图3
基于Magnets的光控诱导重组系统 A:Magnets-PA-Cre和Magnets-PA-Cre3.0系统。蓝光照射下,nMag和pMag的二聚化重构分割型Cre重组酶活性,促使两个loxP位点的目标基因(GOI)发生重组。B:TamPA-Cre系统。通过将胞质定位的雌激素受体(ER)融合到分割型CreN-nMag的N端,使TamPA-Cre蛋白ER-CreN-nMag与核定位的NLS-pMag-CreC在空间上分离。在他莫昔芬Tam处理和蓝光刺激下,分割型Cre重组酶随着nMag-pMag的二聚化而互补重构。C:TRE-PA-Cre系统。tTA依赖的TRE启动子驱动CreN-nMag和CreC-pMag的表达,在没有Dox情况下,蓝光照射激活nMag-pMag二聚化重构分割型Cre重组酶恢复催化活性。"
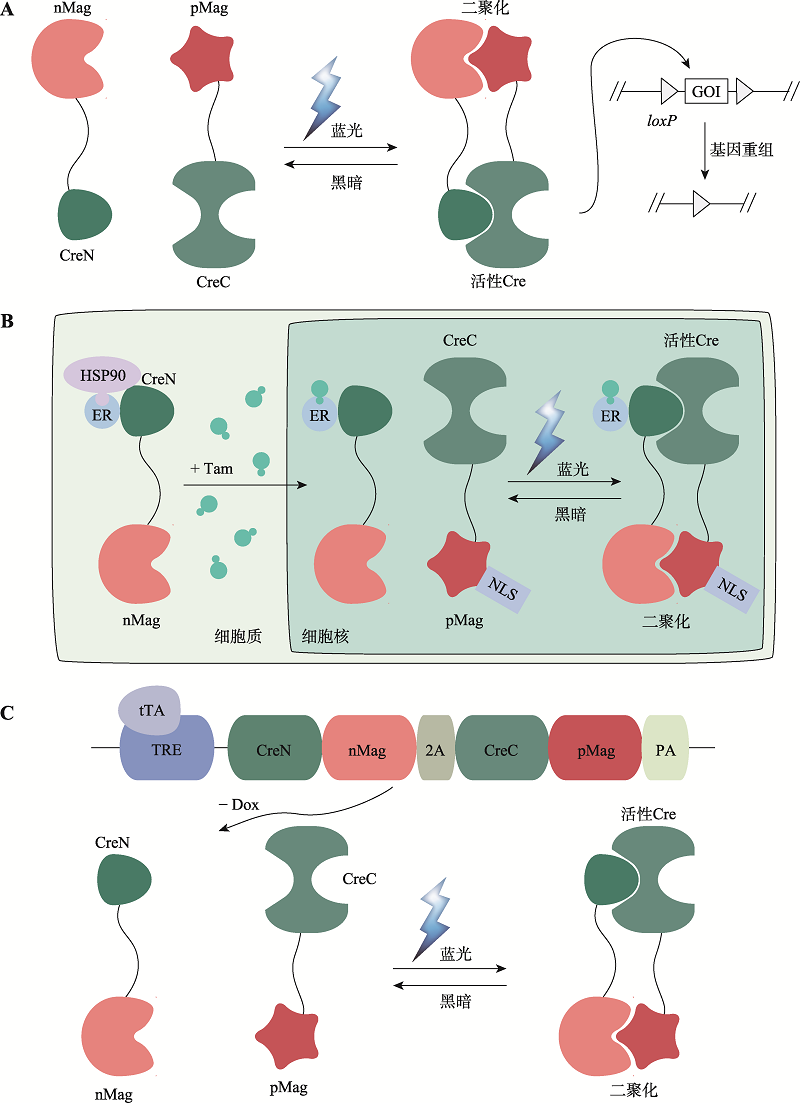
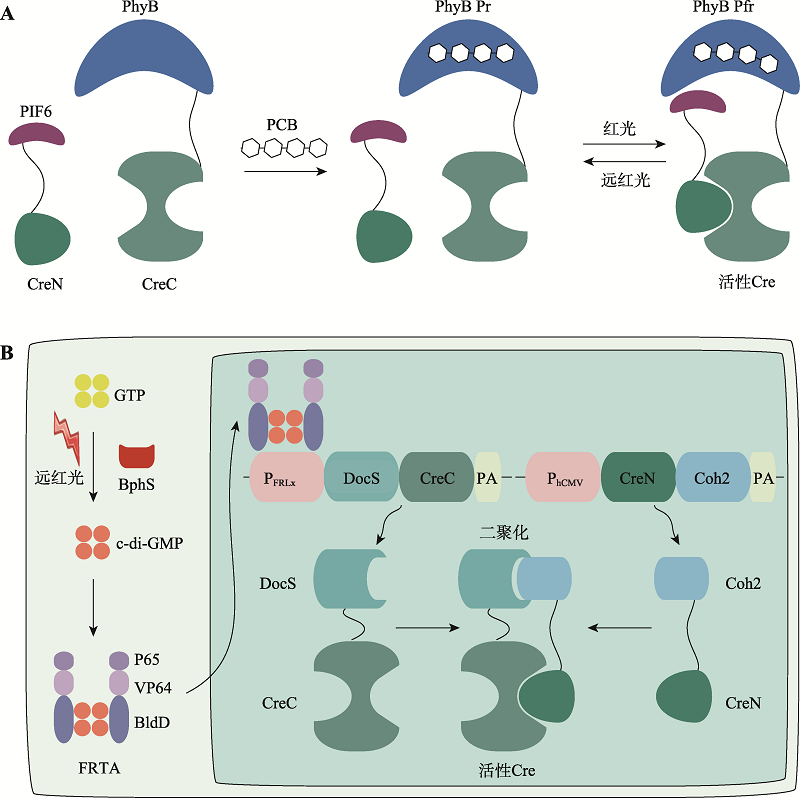
图4
基于光敏色素的光控诱导重组系统 A:基于PhyB的CreLite系统。在这个系统中,PhyB和PIF6分别与CreC和CreN融合。PhyB需要辅助因子PCB才能发挥功能。PhyB与PCB共价结合后吸收红光和红外光。当红光暴露后,PhyB发生构象变化,从失活的Pr形式(红色吸收)转变为有活性的Pfr形式(远红色吸收)。这个过程可以被远红外光逆转。Pfr状态下的PhyB和PIF6相互结合,将分割型Cre重组酶的两个片段组合重构,恢复其重组酶活性。B:基于BphS的FISC系统。在这个系统中,Cre重组酶被分为两个片段,其中CreN与Coh2融合,由组成型启动子PhCMV驱动,CreC与DocS融合,由远红光诱导启动子PFRLx驱动。远红光照射下,光感受器BphS将三磷酸鸟苷酸GTP转化为环二鸟苷酸单磷酸盐c-di-GMP,诱导远红光依赖的转录激活因子FRTA(P65-VP64-BldD)与启动子PFRLx结合,驱动DocS-CreC表达。基于Coh2和DocS结构域的强大亲和力,两个分割型Cre片段组装在一起,恢复Cre重组酶的催化活性。"
| [1] |
Sternberg N, Hamilton D. Bacteriophage P 1 site- specific recombination. I. Recombination between loxP sites. J Mol Biol, 1981, 150(4): 467-486.
pmid: 6276557 |
| [2] |
Sternberg N, Hamilton D, Hoess R. Bacteriophage P 1 site-specific recombination. II. Recombination between loxP and the bacterial chromosome. J Mol Biol, 1981, 150(4): 487-507.
pmid: 6276558 |
| [3] |
Sternberg N. Bacteriophage P 1 site-specific recombination. III. Strand exchange during recombination at lox sites. J Mol Biol, 1981, 150(4): 603-608.
pmid: 6460113 |
| [4] |
Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell, 1989, 59(3): 499-509.
pmid: 2509077 |
| [5] |
Anastassiadis K, Fu J, Patsch C, Hu SB, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis Model Mech, 2009, 2(9-10): 508-515.
doi: 10.1242/dmm.003087 pmid: 19692579 |
| [6] |
Han XM, Zhang ZQ, He LJ, Zhu H, Li Y, Pu WJ, Han MY, Zhao H, Liu K, Li Y, Huang XZ, Zhang MJ, Jin HW, Lv Z, Tang J, Wang JJ, Sun RL, Fei J, Tian XY, Duan SZ, Wang QD, Wang LX, He B, Zhou B. A suite of new Dre recombinase drivers markedly expands the ability to perform intersectional genetic targeting. Cell Stem Cell, 2021, 28(6): 1160-1176.e7.
doi: 10.1016/j.stem.2021.01.007 |
| [7] |
Tronche F, Casanova E, Turiault M, Sahly I, Kellendonk C. When reverse genetics meets physiology: the use of site-specific recombinases in mice. FEBS Lett, 2002, 529(1): 116-121.
doi: 10.1016/S0014-5793(02)03266-0 |
| [8] | Sauer B, Henderson N.Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA, 1988, 85(14): 5166-5170. |
| [9] |
Brault V, Besson V, Magnol L, Duchon A, Hérault Y. Cre/loxP-mediated chromosome engineering of the mouse genome. Handb Exp Pharmacol, 2007(178): 29-48.
pmid: 17203650 |
| [10] |
Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res, 1999, 27(22): 4324-4327.
pmid: 10536138 |
| [11] |
Schönig K, Schwenk F, Rajewsky K, Bujard H. Stringent doxycycline dependent control of CRE recombinase in vivo. Nucleic Acids Res, 2002, 30(23): e134.
doi: 10.1093/nar/gnf134 pmid: 12466566 |
| [12] |
Utomo AR, Nikitin AY, Lee WH. Temporal, spatial, and cell type-specific control of Cre-mediated DNA recombination in transgenic mice. Nat Biotechnol, 1999, 17(11): 1091-1096.
pmid: 10545915 |
| [13] |
Jullien N, Sampieri F, Enjalbert A, Herman JP. Regulation of Cre recombinase by ligand-induced complementation of inactive fragments. Nucleic Acids Res, 2003, 31(21): e131.
doi: 10.1093/nar/gng131 |
| [14] |
Sando R, Baumgaertel K, Pieraut S, Torabi-Rander N, Wandless TJ, Mayford M, Maximov A. Inducible control of gene expression with destabilized Cre. Nat Methods, 2013, 10(11): 1085-1088.
doi: 10.1038/nmeth.2640 |
| [15] |
Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA, 1995, 92(15): 6991-6995.
doi: 10.1073/pnas.92.15.6991 |
| [16] |
Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun, 1997, 237(3): 752-757.
doi: 10.1006/bbrc.1997.7124 |
| [17] |
Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science, 1995, 268(5218): 1766-1769.
pmid: 7792603 |
| [18] |
Hirrlinger J, Scheller A, Hirrlinger PG, Kellert B, Tang WN, Wehr MC, Goebbels S, Reichenbach A, Sprengel R, Rossner MJ, Kirchhoff F. Split-cre complementation indicates coincident activity of different genes in vivo. PLoS One, 2009, 4(1): e4286.
doi: 10.1371/journal.pone.0004286 |
| [19] | Tang JCY, Rudolph S, Dhande OS, Abraira VE, Choi S, Lapan SW, Drew IR, Drokhlyansky E, Huberman AD, Regehr WG, Cepko CL. Cell type-specific manipulation with GFP-dependent Cre recombinase. Nat Neurosci, 2015, 18(9): 1334-1341. |
| [20] | Gengenbacher M, Zimmerman MD, Sarathy JP, Kaya F, Wang H, Mina M, Carter C, Hossen MA, Su HW, Trujillo C, Ehrt S, Schnappinger D, Dartois V. Tissue distribution of doxycycline in animal models of tuberculosis. Antimicrob Agents Chemother, 2020, 64(5): e02479-e02519. |
| [21] |
Wilking N, Appelgren LE, Carlström K, Pousette A, Theve NO. The distribution and metabolism of 14C-labelled tamoxifen in spayed female mice. Acta Pharmacol Toxicol (Copenh), 1982, 50(3): 161-168.
pmid: 7090841 |
| [22] | Zhang JS, Wong SHD, Wu X, Lei H, Qin M, Shi P, Wang W, Bian LM, Cao Y. Engineering photoresponsive ligand tethers for mechanical regulation of stem cells. Adv Mater, 2021, 33(48): e2105765. |
| [23] | Richter F, Fonfara I, Bouazza B, Schumacher CH, Bratovič M, Charpentier E, Möglich A. Engineering of temperature- and light-switchable Cas9 variants. Nucleic Acids Res, 2016, 44(20): 10003-10014. |
| [24] |
Liu RM, Yang J, Yao J, Zhao Z, He W, Su N, Zhang ZY, Zhang CX, Zhang Z, Cai HB, Zhu LY, Zhao YZ, Quan S, Chen XJ, Yang Y. Optogenetic control of RNA function and metabolism using engineered light-switchable RNA-binding proteins. Nat Biotechnol, 2022, 40(5): 779-786.
doi: 10.1038/s41587-021-01112-1 |
| [25] |
Mayer G, Heckel A. Biologically active molecules with a “light switch”. Angew Chem Int Ed Engl, 2006, 45(30): 4900-4921.
doi: 10.1002/anie.200600387 |
| [26] |
Pinheiro AV, Baptista P, Lima JC. Light activation of transcription: photocaging of nucleotides for control over RNA polymerization. Nucleic Acids Res, 2008, 36(14): e90.
doi: 10.1093/nar/gkn415 |
| [27] |
Karginov AV, Zou Y, Shirvanyants D, Kota P, Dokholyan NV, Young DD, Hahn KM, Deiters A. Light regulation of protein dimerization and kinase activity in living cells using photocaged rapamycin and engineered FKBP. J Am Chem Soc, 2011, 133(3): 420-423.
doi: 10.1021/ja109630v pmid: 21162531 |
| [28] |
Miller DS, Chirayil S, Ball HL, Luebke KJ. Manipulating cell migration and proliferation with a light-activated polypeptide. Chembiochem, 2009, 10(3): 577-584.
doi: 10.1002/cbic.200800679 |
| [29] |
Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Control of protein phosphorylation with a genetically encoded photocaged amino acid. Nat Chem Biol, 2007, 3(12): 769-772.
doi: 10.1038/nchembio.2007.44 |
| [30] |
Link KH, Shi YH, Koh JT. Light activated recombination. J Am Chem Soc, 2005, 127(38): 13088-13089.
doi: 10.1021/ja0531226 |
| [31] |
Lawrence DS. The preparation and in vivo applications of caged peptides and proteins. Curr Opin Chem Biol, 2005, 9(6): 570-575.
pmid: 16182597 |
| [32] |
Young DD, Deiters A. Photochemical control of biological processes. Org Biomol Chem, 2007, 5(7): 999-1005.
doi: 10.1039/B616410M |
| [33] |
Tang XJ, Dmochowski IJ. Regulating gene expression with light-activated oligonucleotides. Mol BioSyst, 2007, 3(2): 100-110.
doi: 10.1039/B614349K |
| [34] |
Sinha DK, Neveu P, Gagey N, Aujard I, Benbrahim- Bouzidi C, Le Saux T, Rampon C, Gauron C, Goetz B, Dubruille S, Baaden M, Volovitch M, Bensimon D, Vriz S, Jullien L. Photocontrol of protein activity in cultured cells and zebrafish with one- and two-photon illumination. Chembiochem, 2010, 11(5): 653-663.
doi: 10.1002/cbic.201000008 |
| [35] |
Sinha DK, Neveu P, Gagey N, Aujard I, Le Saux T, Rampon C, Gauron C, Kawakami K, Leucht C, Bally- Cuif L, Volovitch M, Bensimon D, Jullien L, Vriz S. Photoactivation of the CreER T2 recombinase for conditional site-specific recombination with high spatiotemporal resolution. Zebrafish, 2010, 7(2): 199-204.
doi: 10.1089/zeb.2009.0632 |
| [36] |
Lu X, Agasti SS, Vinegoni C, Waterman P, DePinho RA, Weissleder R. Optochemogenetics (OCG) allows more precise control of genetic engineering in mice with CreER regulators. Bioconjug Chem, 2012, 23(9): 1945-1951.
doi: 10.1021/bc300319c |
| [37] |
Inlay MA, Choe V, Bharathi S, Fernhoff NB, Baker JR Jr, Weissman IL, Choi SK. Synthesis of a photocaged tamoxifen for light-dependent activation of Cre-ER recombinase-driven gene modification. Chem Commun (Camb), 2013, 49(43): 4971-4973.
doi: 10.1039/c3cc42179a |
| [38] |
Faal T, Wong PT, Tang SZ, Coulter A, Chen Y, Tu CH, Baker JR, Choi SK, Inlay MA. 4-Hydroxytamoxifen probes for light-dependent spatiotemporal control of Cre-ER mediated reporter gene expression. Mol Biosyst, 2015, 11(3): 783-790.
doi: 10.1039/C4MB00581C |
| [39] |
Brown KA, Zou Y, Shirvanyants D, Zhang J, Samanta S, Mantravadi PK, Dokholyan NV, Deiters A. Light- cleavable rapamycin dimer as an optical trigger for protein dimerization. Chem Commun (Camb), 2015, 51(26): 5702-5705.
doi: 10.1039/C4CC09442E |
| [40] |
Gorka AP, Nani RR, Zhu JJ, Mackem S, Schnermann MJ. A near-IR uncaging strategy based on cyanine photochemistry. J Am Chem Soc, 2014, 136(40): 14153-14159.
doi: 10.1021/ja5065203 |
| [41] |
Edwards WF, Young DD, Deiters A. Light-activated Cre recombinase as a tool for the spatial and temporal control of gene function in mammalian cells. ACS Chem Biol, 2009, 4(6): 441-445.
doi: 10.1021/cb900041s |
| [42] |
Luo J, Arbely E, Zhang J, Chou C, Uprety R, Chin JW, Deiters A. Genetically encoded optical activation of DNA recombination in human cells. Chem Commun (Camb), 2016, 52(55): 8529-8532.
doi: 10.1039/C6CC03934K |
| [43] |
Beverage JN, Sissung TM, Sion AM, Danesi R, Figg WD. CYP2D6 polymorphisms and the impact on tamoxifen therapy. J Pharm Sci, 2007, 96(9): 2224-2231.
doi: 10.1002/jps.20892 |
| [44] |
Cambridge SB, Geissler D, Keller S, Cürten B. A caged doxycycline analogue for photoactivated gene expression. Angew Chem Int Ed Engl, 2006, 45(14): 2229-2231.
doi: 10.1002/anie.200503339 |
| [45] |
Goegan B, Terzi F, Bolze F, Cambridge S, Specht A. Synthesis and characterization of photoactivatable doxycycline analogues bearing two-photon-sensitive photoremovable groups suitable for light-induced gene expression. Chembiochem, 2018, 19(12): 1341-1348.
doi: 10.1002/cbic.201700628 |
| [46] |
Cambridge SB, Geissler D, Calegari F, Anastassiadis K, Hasan MT, Stewart AF, Huttner WB, Hagen V, Bonhoeffer T. Doxycycline-dependent photoactivated gene expression in eukaryotic systems. Nat Methods, 2009, 6(7): 527-531.
doi: 10.1038/nmeth.1340 pmid: 19503080 |
| [47] |
Zheng YJ, Nandakumar KS, Cheng K. Optimization of CAR-T cell-based therapies using small-molecule-based safety switches. J Med Chem, 2021, 64(14): 9577-9591.
doi: 10.1021/acs.jmedchem.0c02054 |
| [48] |
Deiters A, Groff D, Ryu Y, Xie JM, Schultz PG. A genetically encoded photocaged tyrosine. Angew Chem Int Ed Engl, 2006, 45(17): 2728-2731.
doi: 10.1002/anie.200600264 |
| [49] |
Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods, 2010, 7(12): 973-975.
doi: 10.1038/nmeth.1524 |
| [50] |
Li F, Lu ZW, Wu WB, Qian NN, Wang FC, Chen T. Optogenetic gene editing in regional skin. Cell Res, 2019, 29(10): 862-865.
doi: 10.1038/s41422-019-0209-9 |
| [51] | Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods, 2013, 10(3): 249-252. |
| [52] |
Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL. An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun, 2014, 5: 4925.
doi: 10.1038/ncomms5925 pmid: 25233328 |
| [53] |
Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Conformational switching in the fungal light sensor Vivid. Science, 2007, 316(5827): 1054-1057.
pmid: 17510367 |
| [54] |
Zoltowski BD, Crane BR. Light activation of the LOV protein vivid generates a rapidly exchanging dimer. Biochemistry, 2008, 47(27): 7012-7019.
doi: 10.1021/bi8007017 pmid: 18553928 |
| [55] | Vaidya AT, Chen CH, Dunlap JC, Loros JJ, Crane BR. Structure of a light-activated LOV protein dimer that regulates transcription. Sci Signal, 2011, 4(184): ra50. |
| [56] |
Kawano F, Suzuki H, Furuya A, Sato M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun, 2015, 6: 6256.
doi: 10.1038/ncomms7256 pmid: 25708714 |
| [57] |
Liu HT, Yu XH, Li KW, Klejnot J, Yang HY, Lisiero D, Lin CT. Photoexcited CRY 2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science, 2008, 322(5907): 1535-1539.
doi: 10.1126/science.1163927 |
| [58] |
Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotechnol, 2002, 20(10): 1041-1044.
pmid: 12219076 |
| [59] |
Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature, 2009, 461(7266): 997-1001.
doi: 10.1038/nature08446 |
| [60] |
Wu D, Hu Q, Yan Z, Chen W, Yan CY, Huang X, Zhang J, Yang PY, Deng HT, Wang JW, Deng XW, Shi YG.Structural basis of ultraviolet-B perception by UVR8. Nature, 2012, 484(7393): 214-219.
doi: 10.1038/nature10931 |
| [61] |
Cloix C, Kaiserli E, Heilmann M, Baxter KJ, Brown BA, O'Hara A, Smith BO, Christie JM, Jenkins GI. C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein. Proc Natl Acad Sci USA, 2012, 109(40): 16366-16370.
doi: 10.1073/pnas.1210898109 |
| [62] |
Crefcoeur RP, Yin RH, Ulm R, Halazonetis TD. Ultraviolet-B-mediated induction of protein-protein interactions in mammalian cells. Nat Commun, 2013, 4: 1779.
doi: 10.1038/ncomms2800 pmid: 23653191 |
| [63] |
Kaberniuk AA, Shemetov AA, Verkhusha VV. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods, 2016, 13(7): 591-597.
doi: 10.1038/nmeth.3864 pmid: 27159085 |
| [64] |
Ryu MH, Gomelsky M. Near-infrared light responsive synthetic c-di-GMP module for optogenetic applications. ACS Synth Biol, 2014, 3(11): 802-810.
doi: 10.1021/sb400182x |
| [65] |
Shao JW, Xue S, Yu GL, Yu YH, Yang XP, Bai Y, Zhu SC, Yang LF, Yin JL, Wang YD, Liao SY, Guo SW, Xie MQ, Fussenegger M, Ye HF. Smartphone-controlled optogenetically engineered cells enable semiautomatic glucose homeostasis in diabetic mice. Sci Transl Med, 2017, 9(387): eaal2298.
doi: 10.1126/scitranslmed.aal2298 |
| [66] |
Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods, 2012, 9(4): 379-384.
doi: 10.1038/nmeth.1904 pmid: 22388287 |
| [67] |
Woolley GA. Designing chimeric LOV photoswitches. Chem Biol, 2012, 19(4): 441-442.
doi: 10.1016/j.chembiol.2012.04.003 |
| [68] |
Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci USA, 2015, 112(1): 112-117.
doi: 10.1073/pnas.1417910112 |
| [69] |
Wan S, Parrish JA, Anderson RR, Madden M. Transmittance of nonionizing radiation in human tissues. Photochem Photobiol, 1981, 34(6): 679-681.
pmid: 6458827 |
| [70] |
Gomelsky M. Photoactivated cells link diagnosis and therapy. Sci Transl Med, 2017, 9(387): eaan3936.
doi: 10.1126/scitranslmed.aan3936 |
| [71] |
Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, Tucker CL. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat Chem Biol, 2016, 12(6): 425-430.
doi: 10.1038/nchembio.2063 pmid: 27065233 |
| [72] |
Meador K, Wysoczynski CL, Norris AJ, Aoto J, Bruchas MR, Tucker CL. Achieving tight control of a photoactivatable Cre recombinase gene switch: new design strategies and functional characterization in mammalian cells and rodent. Nucleic Acids Res, 2019, 47(17): e97.
doi: 10.1093/nar/gkz585 |
| [73] |
Kawano F, Okazaki R, Yazawa M, Sato M. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat Chem Biol, 2016, 12(12): 1059-1064.
doi: 10.1038/nchembio.2205 pmid: 27723747 |
| [74] |
Morikawa K, Furuhashi K, de Sena-Tomas C, Garcia-Garcia AL, Bekdash R, Klein AD, Gallerani N, Yamamoto HE, Park SHE, Collins GS, Kawano F, Sato M, Lin CS, Targoff KL, Au E, Salling MC, Yazawa M. Photoactivatable Cre recombinase 3.0 for in vivo mouse applications. Nat Commun, 2020, 11(1): 2141.
doi: 10.1038/s41467-020-16030-0 pmid: 32358538 |
| [75] |
Allen ME, Zhou W, Thangaraj J, Kyriakakis P, Wu YQ, Huang ZL, Ho P, Pan YJ, Limsakul P, Xu XD, Wang YX. An AND-Gated drug and photoactivatable Cre-loxP system for spatiotemporal control in cell-based therapeutics. ACS Synth Biol, 2019, 8(10): 2359-2371.
doi: 10.1021/acssynbio.9b00175 |
| [76] |
Takao T, Hiraoka Y, Kawabe K, Yamada D, Ming L, Tanaka K, Sato M, Takarada T. Establishment of a tTA-dependent photoactivatable Cre recombinase knock-in mouse model for optogenetic genome engineering. Biochem Biophys Res Commun, 2020, 526(1): 213-217.
doi: 10.1016/j.bbrc.2020.03.015 |
| [77] |
Yen ST, Trimmer KA, Aboul-Fettouh N, Mullen RD, Culver JC, Dickinson ME, Behringer RR, Eisenhoffer GT. CreLite: An optogenetically controlled Cre/loxP system using red light. Dev Dyn, 2020, 249(11): 1394-1403.
doi: 10.1002/dvdy.232 |
| [78] |
Wu JL, Wang MY, Yang XP, Yi CW, Jiang J, Yu YH, Ye HF. A non-invasive far-red light-induced split-Cre recombinase system for controllable genome engineering in mice. Nat Commun, 2020, 11(1): 3708.
doi: 10.1038/s41467-020-17530-9 |
| [79] |
Jung H, Kim SW, Kim M, Hong J, Yu D, Kim JH, Lee Y, Kim S, Woo D, Shin HS, Park BO, Heo WD. Noninvasive optical activation of Flp recombinase for genetic manipulation in deep mouse brain regions. Nat Commun, 2019, 10(1): 314.
doi: 10.1038/s41467-018-08282-8 |
| [80] |
Li HY, Zhang QS, Gu YR, Wu YY, Wang YM, Wang LR, Feng SJ, Hu YQ, Zheng YS, Li YM, Ye HF, Zhou B, Lin LN, Liu MY, Yang HY, Li DL. Efficient photoactivatable Dre recombinase for cell type-specific spatiotemporal control of genome engineering in the mouse. Proc Natl Acad Sci USA, 2020, 117(52): 33426-33435.
doi: 10.1073/pnas.2003991117 |
| [81] |
Duan LT, Che D, Zhang K, Ong QX, Guo SL, Cui BX. Optogenetic control of molecular motors and organelle distributions in cells. Chem Biol, 2015, 22(5): 671-682.
doi: 10.1016/j.chembiol.2015.04.014 |
| [82] |
Hughes RM, Bolger S, Tapadia H, Tucker CL. Light-mediated control of DNA transcription in yeast. Methods, 2012, 58(4): 385-391.
doi: 10.1016/j.ymeth.2012.08.004 |
| [83] |
Konermann S, Brigham MD, Trevino A, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature, 2013, 500(7463): 472-476.
doi: 10.1038/nature12466 |
| [84] |
Polstein LR, Gersbach CA. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol, 2015, 11(3): 198-200.
doi: 10.1038/nchembio.1753 pmid: 25664691 |
| [85] |
Nihongaki Y, Furuhata Y, Otabe T, Hasegawa S, Yoshimoto K, Sato M. CRISPR-Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat Methods, 2017, 14(10): 963-966.
doi: 10.1038/nmeth.4430 pmid: 28892089 |
| [86] | Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci USA, 2012, 109(35): E2316-E2323. |
| [87] |
Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell, 2013, 153(7): 1494-1509.
doi: 10.1016/j.cell.2013.05.026 |
| [88] |
Kakumoto T, Nakata T. Optogenetic control of PIP3: PIP3 is sufficient to induce the actin-based active part of growth cones and is regulated via endocytosis. PLoS One, 2013, 8(8): e70861.
doi: 10.1371/journal.pone.0070861 |
| [89] |
Maiuri P, Rupprecht JF, Wieser S, Ruprecht V, Bénichou O, Carpi N, Coppey M, De Beco S, Gov N, Heisenberg CP, Lage Crespo C, Lautenschlaeger F, Le Berre M, Lennon-Dumenil AM, Raab M, Thiam HR, Piel M, Sixt M, Voituriez R. Actin flows mediate a universal coupling between cell speed and cell persistence. Cell, 2015, 161(2): 374-386.
doi: 10.1016/j.cell.2015.01.056 pmid: 25799384 |
| [90] |
Boulina M, Samarajeewa H, Baker JD, Kim MD, Chiba A. Live imaging of multicolor-labeled cells in Drosophila. Development, 2013, 140(7): 1605-1613.
doi: 10.1242/dev.088930 pmid: 23482495 |
| [91] |
Schindler SE, McCall JG, Yan P, Hyrc KL, Li MJ, Tucker CL, Lee JM, Bruchas MR, Diamond MI. Photo-activatable Cre recombinase regulates gene expression in vivo. Sci Rep, 2015, 5: 13627.
doi: 10.1038/srep13627 pmid: 26350769 |
| [92] |
Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol, 2009, 5(11): 827-834.
doi: 10.1038/nchembio.210 pmid: 19718042 |
| [93] |
Lamb JS, Zoltowski BD, Pabit SA, Crane BR, Pollack L. Time-resolved dimerization of a PAS-LOV protein measured with photocoupled small angle X-ray scattering. J Am Chem Soc, 2008, 130(37): 12226-12227.
doi: 10.1021/ja804236f |
| [94] | Thomson JG, Rucker EB 3rd, Piedrahita JA. Mutational analysis of loxP sites for efficient Cre-mediated insertion into genomic DNA. Genesis, 2003, 36(3): 162-167. |
| [95] |
Oberdoerffer P, Otipoby KL, Maruyama M, Rajewsky K.Unidirectional Cre-mediated genetic inversion in mice using the mutant loxP pair lox66/lox71. Nucleic Acids Res, 2003, 31(22): e140.
doi: 10.1093/nar/gng140 pmid: 14602933 |
| [96] |
Araki K, Okada Y, Araki M, Yamamura KI. Comparative analysis of right element mutant lox sites on recombination efficiency in embryonic stem cells. BMC Biotechnol, 2010, 10: 29.
doi: 10.1186/1472-6750-10-29 pmid: 20356367 |
| [97] |
Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA, 1992, 89(12): 5547-5551.
doi: 10.1073/pnas.89.12.5547 |
| [98] |
Furth PA, St Onge L, Böger H, Gruss P, Gossen M, Kistner A, Bujard H, Hennighausen L. Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc Natl Acad Sci USA, 1994, 91(20): 9302-9306.
doi: 10.1073/pnas.91.20.9302 |
| [99] |
Jorissen HJMM, Quest B, Lindner I, de Marsac NT, Gärtner W. Phytochromes with noncovalently bound chromophores: the ability of apophytochromes to direct tetrapyrrole photoisomerization. Photochem Photobiol, 2002, 75(5): 554-559.
pmid: 12017484 |
| [100] |
Kami C, Mukougawa K, Muramoto T, Yokota A, Shinomura T, Lagarias JC, Kohchi T. Complementation of phytochrome chromophore-deficient Arabidopsis by expression of phycocyanobilin:ferredoxin oxidoreductase. Proc Natl Acad Sci USA, 2004, 101(4): 1099-1104.
doi: 10.1073/pnas.0307615100 |
| [101] |
Zhou Y, Kong DQ, Wang XY, Yu GL, Wu X, Guan NZ, Weber W, Ye HF. A small and highly sensitive red/far- red optogenetic switch for applications in mammals. Nat Biotechnol, 2022, 40(2): 262-272.
doi: 10.1038/s41587-021-01036-w |
| [102] |
Müller K, Engesser R, Timmer J, Nagy F, Zurbriggen MD, Weber W. Synthesis of phycocyanobilin in mammalian cells. Chem Commun (Camb), 2013, 49(79): 8970-8972.
doi: 10.1039/c3cc45065a |
| [103] |
Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell, 2004, 16(11): 3033-3044.
doi: 10.1105/tpc.104.025643 |
| [104] |
Müller K, Engesser R, Metzger S, Schulz S, Kämpf MM, Busacker M, Steinberg T, Tomakidi P, Ehrbar M, Nagy F, Timmer J, Zubriggen MD, Weber W. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res, 2013, 41(7): e77.
doi: 10.1093/nar/gkt002 |
| [105] |
Wiltbank LB, Kehoe DM. Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat Rev Microbiol, 2019, 17(1): 37-50.
doi: 10.1038/s41579-018-0110-4 pmid: 30410070 |
| [106] |
Kojadinovic M, Laugraud A, Vuillet L, Fardoux J, Hannibal L, Adriano JM, Bouyer P, Giraud E, Verméglio A. Dual role for a bacteriophytochrome in the bioenergetic control of Rhodopseudomonas palustris: enhancement of photosystem synthesis and limitation of respiration. Biochim Biophys Acta, 2008, 1777(2): 163-172.
pmid: 17988648 |
| [107] |
Ulijasz AT, Vierstra RD. Phytochrome structure and photochemistry: recent advances toward a complete molecular picture. Curr Opin Plant Biol, 2011, 14(5): 498-506.
doi: 10.1016/j.pbi.2011.06.002 pmid: 21733743 |
| [108] |
Piatkevich KD, Subach FV, Verkhusha VV. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem Soc Rev, 2013, 42(8): 3441-3452.
doi: 10.1039/c3cs35458j pmid: 23361376 |
| [109] |
Tran MTN, Tanaka J, Hamada M, Sugiyama Y, Sakaguchi S, Nakamura M, Takahashi S, Miwa Y. In vivo image analysis using iRFP transgenic mice. Exp Anim, 2014, 63(3): 311-319.
doi: 10.1538/expanim.63.311 |
| [110] |
Shcherbakova DM, Baloban M, Verkhusha VV. Near- infrared fluorescent proteins engineered from bacterial phytochromes. Curr Opin Chem Biol, 2015, 27: 52-63.
doi: 10.1016/j.cbpa.2015.06.005 pmid: 26115447 |
| [111] |
Redchuk TA, Kaberniuk AA, Verkhusha VV. Near-infrared light-controlled systems for gene transcription regulation, protein targeting and spectral multiplexing. Nat Protoc, 2018, 13(5): 1121-1136.
doi: 10.1038/nprot.2018.022 pmid: 29700485 |
| [112] |
Karimova M, Abi-Ghanem J, Berger N, Surendranath V, Pisabarro MT, Buchholz F. Vika/vox, a novel efficient and specific Cre/loxP-like site-specific recombination system. Nucleic Acids Res, 2013, 41(2): e37.
doi: 10.1093/nar/gks1037 |
| [113] |
Karimova M, Splith V, Karpinski J, Pisabarro MT, Buchholz F. Discovery of Nigri/nox and Panto/pox site-specific recombinase systems facilitates advanced genome engineering. Sci Rep, 2016, 6: 30130.
doi: 10.1038/srep30130 pmid: 27444945 |
| [114] | Liu K, Yu W, Tang MX, Tang J, Liu XX, Liu QZ, Li Y, He LJ, Zhang LB, Evans SM, Tian XY, Lui KO, Zhou B. A dual genetic tracing system identifies diverse and dynamic origins of cardiac valve mesenchyme. Development, 2018, 145(18): dev167775. |
| [115] | Plummer NW, Evsyukova IY, Robertson SD, de Marchena J, Tucker CJ, Jensen P. Expanding the power of recombinase-based labeling to uncover cellular diversity. Development, 2015, 142(24): 4385-4393. |
| [116] |
Karimova M, Baker O, Camgoz A, Naumann R, Buchholz F, Anastassiadis K. A single reporter mouse line for Vika, Flp, Dre, and Cre-recombination. Sci Rep, 2018, 8(1): 14453.
doi: 10.1038/s41598-018-32802-7 pmid: 30262904 |
| [117] | Yao SQ, Yuan P, Ouellette B, Zhou T, Mortrud M, Balaram P, Chatterjee S, Wang Y, Daigle TL, Tasic B, Kuang XL, Gong H, Luo QM, Zeng SQ, Curtright A, Dhaka A, Kahan A, Gradinaru V, Chrapkiewicz R, Schnitzer M, Zeng HK, Cetin A. RecV recombinase system for in vivo targeted optogenomic modifications of single cells or cell populations. Nat Methods, 2020, 17(4): 422-429. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
