遗传 ›› 2024, Vol. 46 ›› Issue (10): 760-778.doi: 10.16288/j.yczz.24-153
收稿日期:2024-05-29
修回日期:2024-08-02
出版日期:2024-08-16
发布日期:2024-08-16
通讯作者:
徐一驰,博士,青年研究员,研究方向:计算生物学/发育生物学。E-mail: xuyichi@fudan.edu.cn作者简介:阙亦宸,临床五年制本科生,专业方向:临床医学。E-mail: 20301030069@fudan.edu.cn阙亦宸和刘清泉并列第一作者。
基金资助:
Yichen Que( ), Qingquan Liu(
), Qingquan Liu( ), Yichi Xu(
), Yichi Xu( )
)
Received:2024-05-29
Revised:2024-08-02
Published:2024-08-16
Online:2024-08-16
Supported by:摘要:
阐明人类胚胎发育的分子调控机制一直是科学家们努力攻克的重大生物学问题之一。然而,由于人类胚胎样本稀少、样本解剖困难、器官结构复杂等因素,解析人类胚胎发育机制面临重重挑战。近年来,随着单细胞组学技术的迅猛发展,人们可以在中心法则的各个环节,系统地解析细胞分化过程中的动态变化,并在空间维度上进行观测与研究,加快了构建人类发育细胞图谱的进程,最终实现对人类发育的细胞分类体系、命运轨迹和三维动态变化的刻画。本文首先介绍了绘制人类发育细胞图谱所使用的单细胞组学技术,然后总结了胚胎发育图谱的最新进展并提出了该领域目前面临的主要问题和挑战,最后对如何利用人类胚胎发育图谱解决生物学和医学的关键问题进行了探讨,以期为利用单细胞组学技术构建并应用人类发育细胞蓝图提供更好的指导。
阙亦宸, 刘清泉, 徐一驰. 人类发育细胞蓝图的现状和挑战[J]. 遗传, 2024, 46(10): 760-778.
Yichen Que, Qingquan Liu, Yichi Xu. Progress and challenges in human developmental cell atlas[J]. Hereditas(Beijing), 2024, 46(10): 760-778.
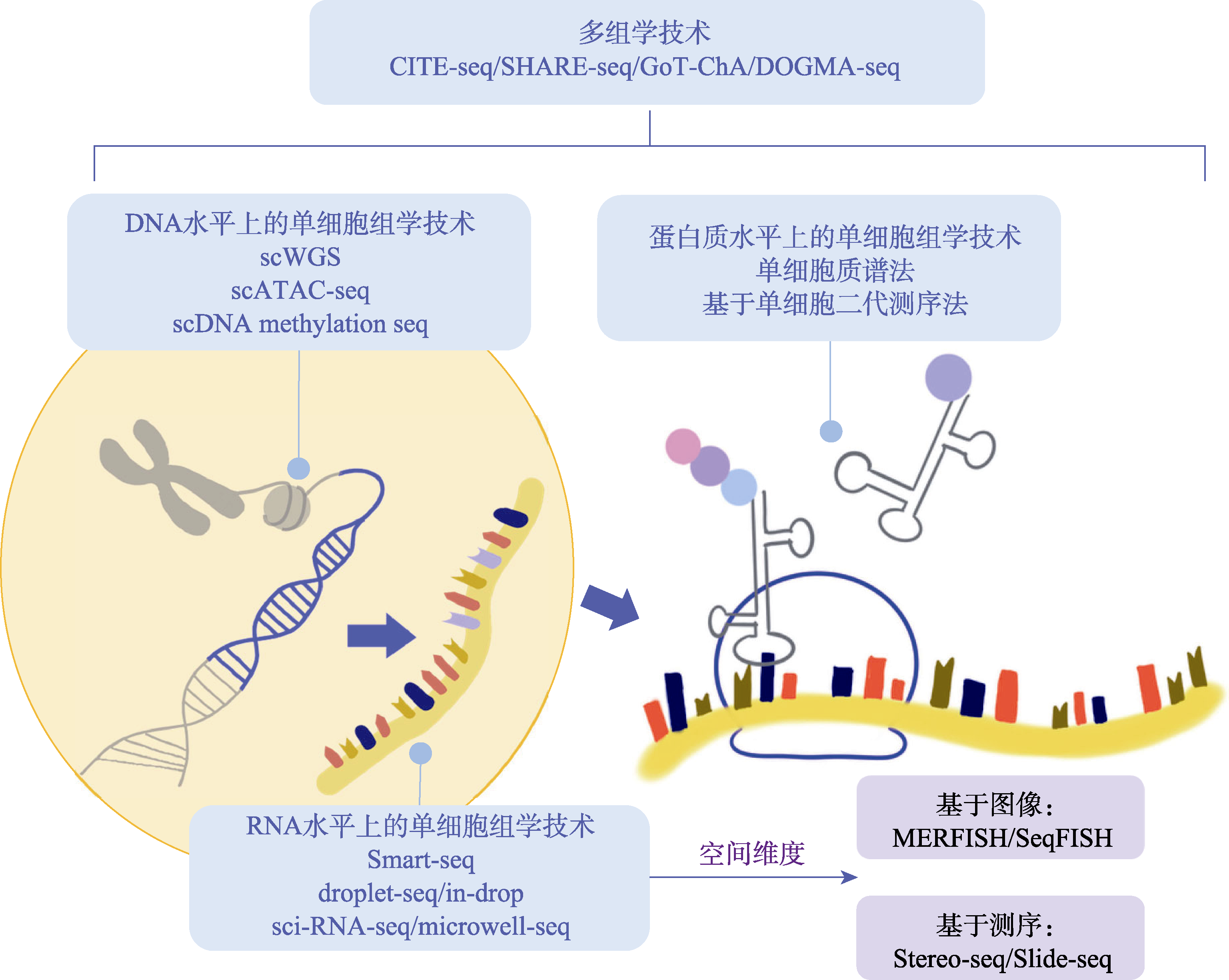
图1
用于人类胚胎发育图谱绘制的单细胞组学技术概览 研究单细胞组学的方法以及空间转录组的各项技术按照中心法则的顺序呈现。CITE-seq:单细胞表面蛋白和转录组测序;SHARE-seq:同步高通量ATAC和RNA表达测序;GoT-ChA:目标区域的单细胞染色质可及性测序;DOGMA-seq:中心法则测序;scWGS-seq:单细胞全基因组测序;scATAC-seq:单细胞染色质可及性测序;scDNA methylation seq:单细胞DNA甲基化测序;Smart-seq:5′端切换的RNA测序;droplet-seq/in-drop:基于微液滴的scRNA-seq;sci-RNA-seq:单细胞组合索引RNA测序;microwell-seq:微孔测序;MERFISH:多重抗差荧光原位杂交;SeqFISH:连续荧光原位杂交;Stereo-seq:空间增强分辨率组学测序;Slide-seq:薄片测序。"
| [1] | Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature, 2001, 409(6822): 860-921. |
| [2] |
Tang FC, Barbacioru C, Wang YZ, Nordman E, Lee C, Xu NL, Wang XH, Bodeau J, Tuch BB, Siddiqui A, Lao KQ, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods, 2009, 6(5): 377-382.
doi: 10.1038/nmeth.1315 pmid: 19349980 |
| [3] |
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell, 2015, 161(5): 1187-1201.
doi: S0092-8674(15)00500-0 pmid: 26000487 |
| [4] |
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell, 2015, 161(5): 1202-1214.
doi: S0092-8674(15)00549-8 pmid: 26000488 |
| [5] |
Svensson V, Vento-Tormo R, Teichmann SA. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc, 2018, 13(4): 599-604.
doi: 10.1038/nprot.2017.149 pmid: 29494575 |
| [6] | Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, Bodenmiller B, Campbell P, Carninci P, Clatworthy M, Clevers H, Deplancke B, Dunham I, Eberwine J, Eils R, Enard W, Farmer A, Fugger L, Göttgens B, Hacohen N, Haniffa M, Hemberg M, Kim S, Klenerman P, Kriegstein A, Lein E, Linnarsson S, Lundberg E, Lundeberg J, Majumder P, Marioni JC, Merad M, Mhlanga M, Nawijn M, Netea M, Nolan G, Pe'er D, Phillipakis A, Ponting CP, Quake S, Reik W, Rozenblatt-Rosen O, Sanes J, Satija R, Schumacher TN, Shalek A, Shapiro E, Sharma P, Shin JW, Stegle O, Stratton M, Stubbington MJT, Theis FJ, Uhlen M, van Oudenaarden A, Wagner A, Watt F, Weissman J, Wold B, Xavier R, Yosef N, Human cell atlas meeting participants. The human cell atlas. Elife, 2017, 6: e27041. |
| [7] | HuBMAP consortium. The human body at cellular resolution: the NIH human biomolecular atlas program. Nature, 2019, 574(7777): 187-192. |
| [8] |
Sikkema L, Ramírez-Suástegui C, Strobl DC, Gillett TE, Zappia L, Madissoon E, Markov NS, Zaragosi LE, Ji Y, Ansari M, Arguel MJ, Apperloo L, Banchero M, Bécavin C, Berg M, Chichelnitskiy E, Chung MI, Collin A, Gay ACA, Gote-Schniering J, Hooshiar Kashani B, Inecik K, Jain M, Kapellos TS, Kole TM, Leroy S, Mayr CH, Oliver AJ, von Papen M, Peter L, Taylor CJ, Walzthoeni T, Xu C, Bui LT, De Donno C, Dony L, Faiz A, Guo MZ, Gutierrez AJ, Heumos L, Huang N, Ibarra IL, Jackson ND, Kadur Lakshminarasimha Murthy P, Lotfollahi M, Tabib T, Talavera-López C, Travaglini KJ, Wilbrey-Clark A, Worlock KB, Yoshida M, Lung biological network consortium, van den Berge M, Bossé Y, Desai TJ, Eickelberg O, Kaminski N, Krasnow MA, Lafyatis R, Nikolic MZ, Powell JE, Rajagopal J, Rojas M, Rozenblatt-Rosen O, Seibold MA, Sheppard D, Shepherd DP, Sin DD, Timens W, Tsankov AM, Whitsett J, Xu Y, Banovich NE, Barbry P, Duong TE, Falk CS, Meyer KB, Kropski JA, Pe'er D, Schiller HB, Tata PR, Schultze JL, Teichmann SA, Misharin AV, Nawijn MC, Luecken MD, Theis FJ. An integrated cell atlas of the lung in health and disease. Nat Med, 2023, 29(6): 1563-1577.
doi: 10.1038/s41591-023-02327-2 pmid: 37291214 |
| [9] | Haniffa M, Taylor D, Linnarsson S, Aronow BJ, Bader GD, Barker RA, Camara PG, Camp JG, Chédotal A, Copp A, Etchevers HC, Giacobini P, Göttgens B, Guo GJ, Hupalowska A, James KR, Kirby E, Kriegstein A, Lundeberg J, Marioni JC, Meyer KB, Niakan KK, Nilsson M, Olabi B, Pe'er D, Regev A, Rood J, Rozenblatt-Rosen O, Satija R, Teichmann SA, Treutlein B, Vento-Tormo R, Webb S, Human cell atlas developmental biological network. A roadmap for the human developmental cell atlas. Nature, 2021, 597(7875): 196-205. |
| [10] |
O'Rahilly R, Müller F. Developmental stages in human embryos: revised and new measurements. Cells Tissues Organs, 2010, 192(2): 73-84.
doi: 10.1159/000289817 pmid: 20185898 |
| [11] |
Billon V, Sanchez-Luque FJ, Rasmussen J, Bodea GO, Gerhardt DJ, Gerdes P, Cheetham SW, Schauer SN, Ajjikuttira P, Meyer TJ, Layman CE, Nevonen KA, Jansz N, Garcia-Perez JL, Richardson SR, Ewing AD, Carbone L, Faulkner GJ. Somatic retrotransposition in the developing rhesus macaque brain. Genome Res, 2022, 32(7): 1298-1314.
doi: 10.1101/gr.276451.121 pmid: 35728967 |
| [12] | Chen CY, Xing D, Tan LZ, Li H, Zhou GY, Huang L, Xie XS. Single-cell whole-genome analyses by linear amplification via transposon insertion. Science, 2017, 356(6334): 189-194. |
| [13] |
van den Bos H, Bakker B, Taudt A, Guryev V, Colomé-Tatché M, Lansdorp PM, Foijer F, Spierings DCJ. Quantification of aneuploidy in mammalian systems. Methods Mol Biol, 2019, 1896: 159-190.
doi: 10.1007/978-1-4939-8931-7_15 pmid: 30474848 |
| [14] |
Wang Z, Shen XH, Shi QH. Advances in single-cell whole genome sequencing technology and its application in biomedicine. Hereditas(Beijing), 2021, 43(2): 108-117.
doi: 10.16288/j.yczz.20-363 pmid: 33724214 |
| 王卓, 申笑涵, 施奇惠. 单细胞基因组测序技术新进展及其在生物医学中的应用. 遗传, 2021, 43(2): 108-117. | |
| [15] | Buenrostro JD, Wu BJ, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature, 2015, 523(7561): 486-490. |
| [16] | Domcke S, Hill AJ, Daza RM, Cao JY, O'Day DR, Pliner HA, Aldinger KA, Pokholok D, Zhang F, Milbank JH, Zager MA, Glass IA, Steemers FJ, Doherty D, Trapnell C, Cusanovich DA, Shendure J. A human cell atlas of fetal chromatin accessibility. Science, 2020, 370(6518): eaba7612. |
| [17] | He YP, Hariharan M, Gorkin DU, Dickel DE, Luo CY, Castanon RG, Nery JR, Lee AY, Zhao Y, Huang H, Williams BA, Trout D, Amrhein H, Fang RX, Chen HM, Li B, Visel A, Pennacchio LA, Ren B, Ecker JR. Spatiotemporal DNA methylome dynamics of the developing mouse fetus. Nature, 2020, 583(7818): 752-759. |
| [18] | Guo HS, Zhu P, Yan LY, Li R, Hu BQ, Lian Y, Yan J, Ren XL, Lin SL, Li JS, Jin XH, Shi XD, Liu P, Wang XY, Wang W, Wei Y, Li XL, Guo F, Wu XL, Fan XY, Yong J, Wen L, Xie SX, Tang FC, Qiao J. The DNA methylation landscape of human early embryos. Nature, 2014, 511(7511): 606-610. |
| [19] | Cao JY, O'Day DR, Pliner HA, Kingsley PD, Deng M, Daza RM, Zager MA, Aldinger KA, Blecher-Gonen R, Zhang F, Spielmann M, Palis J, Doherty D, Steemers FJ, Glass IA, Trapnell C, Shendure J. A human cell atlas of fetal gene expression. Science, 2020, 370(6518): eaba7721. |
| [20] | Han XP, Zhou ZM, Fei LJ, Sun HY, Wang RY, Chen Y, Chen HD, Wang JJ, Tang HN, Ge WH, Zhou YC, Ye F, Jiang MM, Wu JQ, Xiao YY, Jia XN, Zhang TY, Ma XJ, Zhang Q, Bai XL, Lai SJ, Yu CX, Zhu LJ, Lin R, Gao YC, Wang M, Wu YQ, Zhang JM, Zhan RY, Zhu SY, Hu HL, Wang CC, Chen M, Huang H, Liang TB, Chen JH, Wang WL, Zhang D, Guo G. Construction of a human cell landscape at single-cell level. Nature, 2020, 581(7808): 303-309. |
| [21] |
Yan LY, Yang MY, Guo HS, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng XY, Yan J, Huang J, Li M, Wu XL, Wen L, Lao KQ, Li RQ, Qiao J, Tang FC. Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol, 2013, 20(9): 1131-1139.
doi: 10.1038/nsmb.2660 pmid: 23934149 |
| [22] |
Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Analysis of gene expression in single live neurons. Proc Natl Acad Sci USA, 1992, 89(7): 3010-3014.
doi: 10.1073/pnas.89.7.3010 pmid: 1557406 |
| [23] |
Ramsköld D, Luo SJ, Wang YC, Li RB, Deng QL, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol, 2012, 30(8): 777-782.
pmid: 22820318 |
| [24] |
Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods, 2013, 10(11): 1096-1098.
doi: 10.1038/nmeth.2639 pmid: 24056875 |
| [25] |
Hagemann-Jensen M, Ziegenhain C, Chen P, Ramsköld D, Hendriks GJ, Larsson AJM, Faridani OR, Sandberg R. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat Biotechnol, 2020, 38(6): 708-714.
doi: 10.1038/s41587-020-0497-0 pmid: 32518404 |
| [26] |
Han XP, Wang RY, Zhou YC, Fei LJ, Sun HY, Lai SJ, Saadatpour A, Zhou ZM, Chen HD, Ye F, Huang D, Xu Y, Huang WT, Jiang MM, Jiang XY, Mao J, Chen Y, Lu CY, Xie J, Fang Q, Wang YB, Yue R, Li TF, Huang H, Orkin SH, Yuan GC, Chen M, Guo GJ. Mapping the mouse cell atlas by microwell-seq. Cell, 2018, 172(5): 1091-1107.e17.
doi: S0092-8674(18)30116-8 pmid: 29474909 |
| [27] |
Bennett HM, Stephenson W, Rose CM, Darmanis S. Single-cell proteomics enabled by next-generation sequencing or mass spectrometry. Nat Methods, 2023, 20(3): 363-374.
doi: 10.1038/s41592-023-01791-5 pmid: 36864196 |
| [28] |
Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe'er D, Tanner SD, Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science, 2011, 332(6030): 687-696.
doi: 10.1126/science.1198704 pmid: 21551058 |
| [29] |
Stoeckius M, Hafemeister C, Stephenson W, Houck- Loomis B, Chattopadhyay PK, Swerdlow H, Satija R, Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods, 2017, 14(9): 865-868.
doi: 10.1038/nmeth.4380 pmid: 28759029 |
| [30] |
Ma S, Zhang B, LaFave LM, Earl AS, Chiang Z, Hu Y, Ding JR, Brack A, Kartha VK, Tay T, Law T, Lareau C, Hsu YC, Regev A, Buenrostro JD. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell, 2020, 183(4): 1103-1116.e20.
doi: 10.1016/j.cell.2020.09.056 pmid: 33098772 |
| [31] | Mimitou EP, Lareau CA, Chen KY, Zorzetto-Fernandes AL, Hao YH, Takeshima Y, Luo W, Huang TS, Yeung BZ, Papalexi E, Thakore PI, Kibayashi T, Wing JB, Hata M, Satija R, Nazor KL, Sakaguchi S, Ludwig LS, Sankaran VG, Regev A, Smibert P. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat Biotechnol, 2021, 39(10): 1246-1258. |
| [32] | Izzo F, Myers RM, Ganesan S, Mekerishvili L, Kottapalli S, Prieto T, Eton EO, Botella T, Dunbar AJ, Bowman RL, Sotelo J, Potenski C, Mimitou EP, Stahl M, El Ghaity-Beckley S, Arandela J, Raviram R, Choi DC, Hoffman R, Chaligné R, Abdel-Wahab O, Smibert P, Ghobrial IM, Scandura JM, Marcellino B, Levine RL, Landau DA. Mapping genotypes to chromatin accessibility profiles in single cells. Nature, 2024, 629(8014): 1149-1157. |
| [33] | Rao A, Barkley D, França GS, Yanai I. Exploring tissue architecture using spatial transcriptomics. Nature, 2021, 596(7871): 211-220. |
| [34] | Ke RQ, Mignardi M, Pacureanu A, Svedlund J, Botling J, Wählby C, Nilsson M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods, 2013, 10(9): 857-860. |
| [35] | Moffitt JR, Bambah-Mukku D, Eichhorn SW, Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao JJ, Regev A, Dulac C, Zhuang XW. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science, 2018, 362(6416): eaau5324. |
| [36] | Eng CL, Lawson M, Zhu Q, Dries R, Koulena N, Takei Y, Yun J, Cronin C, Karp C, Yuan GC, Cai L. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature, 2019, 568(7751): 235-239. |
| [37] |
Ståhl PL, Salmén F, Vickovic S, Lundmark A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss M, Mollbrink A, Linnarsson S, Codeluppi S, Borg Å, Pontén F, Costea PI, Sahlén P, Mulder J, Bergmann O, Lundeberg J, Frisén J. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science, 2016, 353(6294): 78-82.
doi: 10.1126/science.aaf2403 pmid: 27365449 |
| [38] |
Chen A, Liao S, Cheng MN, Ma KL, Wu L, Lai YW, Qiu XJ, Yang J, Xu JS, Hao SJ, Wang X, Lu HF, Chen X, Liu X, Huang X, Li Z, Hong Y, Jiang YJ, Peng J, Liu S, Shen MZ, Liu CY, Li QS, Yuan Y, Wei XY, Zheng HW, Feng WM, Wang ZF, Liu Y, Wang ZH, Yang YZ, Xiang HT, Han L, Qin BM, Guo PC, Lai GY, Muñoz-Cánoves P, Maxwell PH, Thiery JP, Wu QF, Zhao FX, Chen BC, Li M, Dai X, Wang S, Kuang HY, Hui JH, Wang LQ, Fei JF, Wang O, Wei XF, Lu HR, Wang B, Liu SP, Gu Y, Ni M, Zhang WW, Mu F, Yin Y, Yang HM, Lisby M, Cornall RJ, Mulder J, Uhlén M, Esteban MA, Li YX, Liu LQ, Xu X, Wang J. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell, 2022, 185(10): 1777-1792.e21.
doi: 10.1016/j.cell.2022.04.003 pmid: 35512705 |
| [39] |
Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F, Macosko EZ. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science, 2019, 363(6434): 1463-1467.
doi: 10.1126/science.aaw1219 pmid: 30923225 |
| [40] | Jiang FQ, Zhou X, Qian YY, Zhu M, Wang L, Li ZX, Shen QM, Wang MH, Qu FF, Cui GZ, Chen K, Peng GD. Simultaneous profiling of spatial gene expression and chromatin accessibility during mouse brain development. Nat Methods, 2023, 20(7): 1048-1057. |
| [41] |
Liu Y, Yang MY, Deng YX, Su G, Enninful A, Guo CC, Tebaldi T, Zhang D, Kim D, Bai ZL, Norris E, Pan A, Li JT, Xiao Y, Halene S, Fan R. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell, 2020, 183(6): 1665-1681.e18.
doi: 10.1016/j.cell.2020.10.026 pmid: 33188776 |
| [42] | Tyser RCV, Mahammadov E, Nakanoh S, Vallier L, Scialdone A, Srinivas S. Single-cell transcriptomic characterization of a gastrulating human embryo. Nature, 2021, 600(7888): 285-289. |
| [43] |
Xiao ZY, Cui L, Yuan Y, He NN, Xie XW, Lin SR, Yang XL, Zhang X, Shi PF, Wei ZF, Li Y, Wang HM, Wang XY, Wei YL, Guo JT, Yu LQ. 3D reconstruction of a gastrulating human embryo. Cell, 2024, 187(11): 2855-2874.e19.
doi: 10.1016/j.cell.2024.03.041 pmid: 38657603 |
| [44] | Zeng B, Liu ZY, Lu YF, Zhong SJ, Qin SY, Huang LW, Zeng Y, Li ZX, Dong H, Shi YC, Yang JL, Dai YL, Ma Q, Sun L, Bian LH, Han D, Chen YQ, Qiu X, Wang W, Marín O, Wu Q, Wang YJ, Wang XQ. The single-cell and spatial transcriptional landscape of human gastrulation and early brain development. Cell Stem Cell, 2023, 30(6): 851-866.e7. |
| [45] |
Xu YC, Zhang TJ, Zhou Q, Hu MZ, Qi Y, Xue YF, Nie YX, Wang LH, Bao ZR, Shi WY. A single-cell transcriptome atlas profiles early organogenesis in human embryos. Nat Cell Biol, 2023, 25(4): 604-615.
doi: 10.1038/s41556-023-01108-w pmid: 36928764 |
| [46] | Farah EN, Hu RK, Kern C, Zhang QQ, Lu TY, Ma QX, Tran S, Zhang B, Carlin D, Monell A, Blair AP, Wang ZL, Eschbach J, Li B, Destici E, Ren B, Evans SM, Chen SC, Zhu Q, Chi NC. Spatially organized cellular communities form the developing human heart. Nature, 2024, 627(8005): 854-864. |
| [47] | Miller AJ, Yu QH, Czerwinski M, Tsai YH, Conway RF, Wu A, Holloway EM, Walker T, Glass IA, Treutlein B, Camp JG, Spence JR. In vitro and in vivo development of the human airway at single-cell resolution. Dev Cell, 2020, 53(1): 117-128.e6. |
| [48] |
He P, Lim K, Sun DW, Pett JP, Jeng Q, Polanski K, Dong ZQ, Bolt L, Richardson L, Mamanova L, Dabrowska M, Wilbrey-Clark A, Madissoon E, Tuong ZK, Dann E, Suo CQ, Goh I, Yoshida M, Nikolić MZ, Janes SM, He XL, Barker RA, Teichmann SA, Marioni JC, Meyer KB, Rawlins EL. A human fetal lung cell atlas uncovers proximal-distal gradients of differentiation and key regulators of epithelial fates. Cell, 2022, 185(25): 4841-4860.e25.
doi: 10.1016/j.cell.2022.11.005 pmid: 36493756 |
| [49] |
Fawkner-Corbett D, Antanaviciute A, Parikh K, Jagielowicz M, Gerós AS, Gupta T, Ashley N, Khamis D, Fowler D, Morrissey E, Cunningham C, Johnson PRV, Koohy H, Simmons A. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell, 2021, 184(3): 810-826.e23.
doi: 10.1016/j.cell.2020.12.016 pmid: 33406409 |
| [50] |
Elmentaite R, Ross ADB, Roberts K, James KR, Ortmann D, Gomes T, Nayak K, Tuck L, Pritchard S, Bayraktar OA, Heuschkel R, Vallier L, Teichmann SA, Zilbauer M. Single-cell sequencing of developing human gut reveals transcriptional links to childhood Crohn's disease. Dev Cell, 2020, 55(6): 771-783.e5.
doi: 10.1016/j.devcel.2020.11.010 pmid: 33290721 |
| [51] | Braun E, Danan-Gotthold M, Borm LE, Lee KW, Vinsland E, Lönnerberg P, Hu LJ, Li XF, He XL, Andrusivova Ž, Lundeberg J, Barker RA, Arenas E, Sundström E, Linnarsson S. Comprehensive cell atlas of the first-trimester developing human brain. Science, 2023, 382(6667): eadf1226. |
| [52] |
Li YX, Li ZQ, Wang CL, Yang M, He ZQ, Wang FY, Zhang YH, Li R, Gong YX, Wang BH, Fan BG, Wang CY, Chen L, Li H, Shi PF, Wang NN, Wei ZF, Wang YL, Jin L, Du P, Dong J, Jiao JW. Spatiotemporal transcriptome atlas reveals the regional specification of the developing human brain. Cell, 2023, 186(26): 5892-5909.e22.
doi: 10.1016/j.cell.2023.11.016 pmid: 38091994 |
| [53] |
Stewart BJ, Ferdinand JR, Young MD, Mitchell TJ, Loudon KW, Riding AM, Richoz N, Frazer GL, Staniforth JUL, Vieira Braga FA, Botting RA, Popescu DM, Vento-Tormo R, Stephenson E, Cagan A, Farndon SJ, Polanski K, Efremova M, Green K, Del Castillo Velasco-Herrera M, Guzzo C, Collord G, Mamanova L, Aho T, Armitage JN, Riddick ACP, Mushtaq I, Farrell S, Rampling D, Nicholson J, Filby A, Burge J, Lisgo S, Lindsay S, Bajenoff M, Warren AY, Stewart GD, Sebire N, Coleman N, Haniffa M, Teichmann SA, Behjati S, Clatworthy MR. Spatiotemporal immune zonation of the human kidney. Science, 2019, 365(6460): 1461-1466.
doi: 10.1126/science.aat5031 pmid: 31604275 |
| [54] | Zhang B, He P, Lawrence JEG, Wang SY, Tuck E, Williams BA, Roberts K, Kleshchevnikov V, Mamanova L, Bolt L, Polanski K, Li T, Elmentaite R, Fasouli ES, Prete M, He XL, Yayon N, Fu YX, Yang H, Liang C, Zhang H, Blain R, Chedotal A, FitzPatrick DR, Firth H, Dean A, Bayraktar OA, Marioni JC, Barker RA, Storer MA, Wold BJ, Zhang HB, Teichmann SA. A human embryonic limb cell atlas resolved in space and time. Nature, 2023. |
| [55] | Zhong SJ, Zhang S, Fan XY, Wu Q, Yan LY, Dong J, Zhang HF, Li L, Sun L, Pan N, Xu XH, Tang FC, Zhang J, Qiao J, Wang XQ. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature, 2018, 555(7697): 524-528. |
| [56] |
Fan XY, Dong J, Zhong SJ, Wei Y, Wu Q, Yan LY, Yong J, Sun L, Wang XY, Zhao YY, Wang W, Yan J, Wang XQ, Qiao J, Tang FC. Spatial transcriptomic survey of human embryonic cerebral cortex by single-cell RNA-seq analysis. Cell Res, 2018, 28(7): 730-745.
doi: 10.1038/s41422-018-0053-3 pmid: 29867213 |
| [57] |
Trevino AE, Müller F, Andersen J, Sundaram L, Kathiria A, Shcherbina A, Farh K, Chang HY, Pasca AM, Kundaje A, Pașca SP, Greenleaf WJ. Chromatin and gene-regulatory dynamics of the developing human cerebral cortex at single-cell resolution. Cell, 2021, 184(19): 5053-5069 e23.
doi: 10.1016/j.cell.2021.07.039 pmid: 34390642 |
| [58] |
Yu Y, Zeng ZW, Xie DL, Chen RL, Sha YQ, Huang SY, Cai WJ, Chen WH, Li WJ, Ke RQ, Sun T. Interneuron origin and molecular diversity in the human fetal brain. Nat Neurosci, 2021, 24(12): 1745-1756.
doi: 10.1038/s41593-021-00940-3 pmid: 34737447 |
| [59] |
Aldinger KA, Thomson Z, Phelps IG, Haldipur P, Deng M, Timms AE, Hirano M, Santpere G, Roco C, Rosenberg AB, Lorente-Galdos B, Gulden FO, O'Day D, Overman LM, Lisgo SN, Alexandre P, Sestan N, Doherty D, Dobyns WB, Seelig G, Glass IA, Millen KJ. Spatial and cell type transcriptional landscape of human cerebellar development. Nat Neurosci, 2021, 24(8): 1163-1175.
doi: 10.1038/s41593-021-00872-y pmid: 34140698 |
| [60] |
Zhang S, Cui YL, Ma XY, Yong J, Yan LY, Yang M, Ren J, Tang FC, Wen L, Qiao J. Single-cell transcriptomics identifies divergent developmental lineage trajectories during human pituitary development. Nat Commun, 2020, 11(1): 5275.
doi: 10.1038/s41467-020-19012-4 pmid: 33077725 |
| [61] | Zhong SJ, Ding WY, Sun L, Lu YF, Dong H, Fan XY, Liu ZY, Chen RG, Zhang S, Ma Q, Tang FC, Wu Q, Wang XQ. Decoding the development of the human hippocampus. Nature, 2020, 577(7791): 531-536. |
| [62] | Herb BR, Glover HJ, Bhaduri A, Colantuoni C, Bale TL, Siletti K, Hodge R, Lein E, Kriegstein AR, Doege CA, Ament SA. Single-cell genomics reveals region-specific developmental trajectories underlying neuronal diversity in the human hypothalamus. Sci Adv, 2023, 9(45): eadf6251. |
| [63] | Popescu DM, Botting RA, Stephenson E, Green K, Webb S, Jardine L, Calderbank EF, Polanski K, Goh I, Efremova M, Acres M, Maunder D, Vegh P, Gitton Y, Park JE, Vento-Tormo R, Miao ZC, Dixon D, Rowell R, McDonald D, Fletcher J, Poyner E, Reynolds G, Mather M, Moldovan C, Mamanova L, Greig F, Young MD, Meyer KB, Lisgo S, Bacardit J, Fuller A, Millar B, Innes B, Lindsay S, Stubbington MJT, Kowalczyk MS, Li B, Ashenberg O, Tabaka M, Dionne D, Tickle TL, Slyper M, Rozenblatt-Rosen O, Filby A, Carey P, Villani AC, Roy A, Regev A, Chédotal A, Roberts I, Göttgens B, Behjati S, Laurenti E, Teichmann SA, Haniffa M. Decoding human fetal liver haematopoiesis. Nature, 2019, 574(7778): 365-371. |
| [64] |
Yu QH, Kilik U, Holloway EM, Tsai YH, Harmel C, Wu A, Wu JH, Czerwinski M, Childs CJ, He ZS, Capeling MM, Huang S, Glass IA, Higgins PDR, Treutlein B, Spence JR, Camp JG. Charting human development using a multi-endodermal organ atlas and organoid models. Cell, 2021, 184(12): 3281-3298.e22.
doi: 10.1016/j.cell.2021.04.028 pmid: 34019796 |
| [65] |
Liu C, Gong YD, Zhang H, Yang H, Zeng Y, Bian ZL, Xin Q, Bai ZJ, Zhang M, He J, Yan J, Zhou J, Li ZC, Ni YL, Wen AQ, Lan Y, Hu HB, Liu B. Delineating spatiotemporal and hierarchical development of human fetal innate lymphoid cells. Cell Res, 2021, 31(10): 1106-1122.
doi: 10.1038/s41422-021-00529-2 pmid: 34239074 |
| [66] |
Zeng Y, Liu C, Gong YD, Bai ZJ, Hou SY, He J, Bian ZL, Li ZC, Ni YL, Yan J, Huang T, Shi H, Ma CY, Chen XY, Wang JY, Bian LH, Lan Y, Liu B, Hu HB. Single-cell RNA sequencing resolves spatiotemporal development of pre-thymic lymphoid progenitors and thymus organogenesis in human embryos. Immunity, 2019, 51(5): 930-948 e6.
doi: S1074-7613(19)30404-2 pmid: 31604687 |
| [67] | Bian ZL, Gong YD, Huang T, Lee CZW, Bian LH, Bai ZJ, Shi H, Zeng Y, Liu C, He J, Zhou J, Li XL, Li ZC, Ni YL, Ma CY, Cui L, Zhang R, Chan JKY, Ng LG, Lan Y, Ginhoux F, Liu B. Deciphering human macrophage development at single-cell resolution. Nature, 2020, 582(7813): 571-576. |
| [68] |
Xu CL, He J, Wang HT, Zhang YN, Wu J, Zhao L, Li Y, Gao J, Geng GF, Wang BR, Chen XY, Zheng ZF, Shen B, Zeng Y, Bai ZJ, Yang H, Shi SJ, Dong F, Ma SH, Jiang E, Cheng T, Lan Y, Zhou JX, Liu B, Shi LH. Single-cell transcriptomic analysis identifies an immune-prone population in erythroid precursors during human ontogenesis. Nat Immunol, 2022, 23(7): 1109-1120.
doi: 10.1038/s41590-022-01245-8 pmid: 35761081 |
| [69] |
Wang HT, He J, Xu CL, Chen XY, Yang H, Shi SJ, Liu CC, Zeng Y, Wu D, Bai ZJ, Wang MG, Wen YQ, Su P, Xia MJ, Huang BM, Ma CY, Bian LH, Lan Y, Cheng T, Shi LH, Liu B, Zhou JX. Decoding human megakaryocyte development. Cell Stem Cell, 2021, 28(3): 535-549.e8.
doi: 10.1016/j.stem.2020.11.006 pmid: 33340451 |
| [70] | Schoenwolf GC, Bleyl SB, Brauer PR, Francis-West PH. Larsen's human embryology. 2008. |
| [71] |
Clifton K, Anant M, Aihara G, Atta L, Aimiuwu OK, Kebschull JM, Miller MI, Tward D, Fan J. STalign: alignment of spatial transcriptomics data using diffeomorphic metric mapping. Nat Commun, 2023, 14(1): 8123.
doi: 10.1038/s41467-023-43915-7 pmid: 38065970 |
| [72] |
Lopez R, Regier J, Cole MB, Jordan MI, Yosef N. Deep generative modeling for single-cell transcriptomics. Nat Methods, 2018, 15(12): 1053-1058.
doi: 10.1038/s41592-018-0229-2 pmid: 30504886 |
| [73] |
Shaham U, Stanton KP, Zhao J, Li HM, Raddassi K, Montgomery R, Kluger Y. Removal of batch effects using distribution-matching residual networks. Bioinformatics, 2017, 33(16): 2539-2546.
doi: 10.1093/bioinformatics/btx196 pmid: 28419223 |
| [74] |
Xu CL, Lopez R, Mehlman E, Regier J, Jordan MI, Yosef N. Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol Syst Biol, 2021, 17(1): e9620.
doi: 10.15252/msb.20209620 pmid: 33491336 |
| [75] |
Sha YT, Qiu YC, Zhou PJ, Nie Q. Reconstructing growth and dynamic trajectories from single-cell transcriptomics data. Nat Mach Intell, 2024, 6(1): 25-39.
doi: 10.1038/s42256-023-00763-w pmid: 38274364 |
| [76] |
Li Q. scTour: a deep learning architecture for robust inference and accurate prediction of cellular dynamics. Genome Biol, 2023, 24(1): 149.
doi: 10.1186/s13059-023-02988-9 pmid: 37353848 |
| [77] | Luecken MD, Büttner M, Chaichoompu K, Danese A, Interlandi M, Mueller MF, Strobl DC, Zappia L, Dugas M, Colomé-Tatché M, Theis FJ. Benchmarking atlas-level data integration in single-cell genomics. Nat Methods, 2022, 19(1): 41-50. |
| [78] |
Arendt D, Musser JM, Baker CVH, Bergman A, Cepko C, Erwin DH, Pavlicev M, Schlosser G, Widder S, Laubichler MD, Wagner GP. The origin and evolution of cell types. Nat Rev Genet, 2016, 17(12): 744-757.
doi: 10.1038/nrg.2016.127 pmid: 27818507 |
| [79] | Greulich P, MacArthur BD, Parigini C, Sanchez-Garcia RJ. Universal principles of lineage architecture and stem cell identity in renewing tissues. Development, 2021, 148(11): dev194399. |
| [80] |
Domcke S, Shendure J. A reference cell tree will serve science better than a reference cell atlas. Cell, 2023, 186(6): 1103-1114.
doi: 10.1016/j.cell.2023.02.016 pmid: 36931241 |
| [81] |
Fleck JS, Camp JG, Treutlein B. What is a cell type? Science, 2023, 381(6659): 733-734.
doi: 10.1126/science.adf6162 pmid: 37590360 |
| [82] |
Shao X, Yang HH, Zhuang X, Liao J, Yang PH, Cheng JY, Lu XY, Chen HJ, Fan XH. scDeepSort: a pre-trained cell-type annotation method for single-cell transcriptomics using deep learning with a weighted graph neural network. Nucleic Acids Res, 2021, 49(21): e122.
doi: 10.1093/nar/gkab775 pmid: 34500471 |
| [83] | Dong XS, Chowdhury S, Victor U, Li XF, Qian LJ. Semi-supervised deep learning for cell type identification from single-cell transcriptomic data. IEEE/ACM Trans Comput Biol Bioinform, 2023, 20(2): 1492-1505. |
| [84] | Chan MM, Smith ZD, Grosswendt S, Kretzmer H, Norman TM, Adamson B, Jost M, Quinn JJ, Yang D, Jones MG, Khodaverdian A, Yosef N, Meissner A, Weissman JS. Molecular recording of mammalian embryogenesis. Nature, 2019, 570(7759): 77-82. |
| [85] | Cao JY, Spielmann M, Qiu XJ, Huang XF, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, Trapnell C, Shendure J. The single-cell transcriptional landscape of mammalian organogenesis. Nature, 2019, 566(7745): 496-502. |
| [86] | Farrell JA, Wang YQ, Riesenfeld SJ, Shekhar K, Regev A, Schier AF. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science, 2018, 360(6392): eaar3131. |
| [87] |
Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science, 2018, 360(6392): 981-987.
doi: 10.1126/science.aar4362 pmid: 29700229 |
| [88] | Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science, 2018, 360(6392): eaar5780. |
| [89] | Zhai JL, Guo J, Wan HF, Qi LQ, Liu LZ, Xiao ZY, Yan L, Schmitz DA, Xu YH, Yu DN, Wu XL, Zhao WT, Yu KY, Jiang XX, Guo F, Wu J, Wang HM. Primate gastrulation and early organogenesis at single-cell resolution. Nature, 2022, 612(7941): 732-738. |
| [90] | Packer JS, Zhu Q, Huynh C, Sivaramakrishnan P, Preston E, Dueck H, Stefanik D, Tan K, Trapnell C, Kim J, Waterston RH, Murray JI. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science, 2019, 365(6459): eaax1971. |
| [91] | Calderon D, Blecher-Gonen R, Huang XF, Secchia S, Kentro J, Daza RM, Martin B, Dulja A, Schaub C, Trapnell C, Larschan E, O'Connor-Giles KM, Furlong EEM, Shendure J. The continuum of Drosophila embryonic development at single-cell resolution. Science, 2022, 377(6606): eabn5800. |
| [92] | Zhang TJ, Xu YC, Imai K, Fei T, Wang GL, Dong B, Yu TW, Satou Y, Shi WY, Bao ZR. A single-cell analysis of the molecular lineage of chordate embryogenesis. Sci Adv, 2020, 6(45): eabc4773. |
| [93] |
Johansen N, Quon G. scAlign: a tool for alignment, integration, and rare cell identification from scRNA-seq data. Genome Biol, 2019, 20(1): 166.
doi: 10.1186/s13059-019-1766-4 pmid: 31412909 |
| [94] | Wang X, Yang L, Wang YC, Xu ZR, Feng Y, Zhang J, Wang Y, Xu CR. Comparative analysis of cell lineage differentiation during hepatogenesis in humans and mice at the single-cell transcriptome level. Cell Res, 2020, 30(12): 1109-1126. |
| [95] | Cao ST, Feng HJ, Yi HY, Pan MJ, Lin LH, Zhang YS, Feng ZY, Liang WF, Cai BM, Li Q, Xiong Z, Shen QM, Ke MJ, Zhao X, Chen HL, He Q, Min MW, Cai QY, Liu H, Wang J, Pei DQ, Chen JK, Ma YL. Single-cell RNA sequencing reveals the developmental program underlying proximal-distal patterning of the human lung at the embryonic stage. Cell Res, 2023, 33(6): 421-433. |
| [96] |
Wagner DE, Klein AM. Lineage tracing meets single-cell omics: opportunities and challenges. Nat Rev Genet, 2020, 21(7): 410-427.
doi: 10.1038/s41576-020-0223-2 pmid: 32235876 |
| [97] | D'Alton ME, DeCherney AH. Prenatal diagnosis. N Engl J Med, 1993, 328(2): 114-20. |
| [98] |
Zhang JL, Wu YT, Chen SC, Luo Q, Xi H, Li JL, Qin XM, Peng Y, Ma N, Yang BX, Qiu X, Lu WL, Chen Y, Jiang Y, Chen PP, Liu YF, Zhang C, Zhang ZW, Xiong Y, Shen J, Liang H, Ren YY, Ying CM, Dong MY, Li XT, Xu CJ, Wang H, Zhang D, Xu CM, Huang HF. Prospective prenatal cell-free DNA screening for genetic conditions of heterogenous etiologies. Nat Med, 2024, 30(2): 470-479.
doi: 10.1038/s41591-023-02774-x pmid: 38253798 |
| [99] |
Qiu XF, Tang DE, Yu HY, Liao QY, Hu ZY, Zhou J, Zhao X, He HY, Liang ZJ, Xu CM, Yang M, Dai Y. Analysis of transcription factors in accessible open chromatin in the 18-trisomy syndrome based on single cell ATAC sequencing technique. Hereditas(Beijing), 2021, 43(1): 74-83.
doi: 10.16288/j.yczz.20-283 pmid: 33509776 |
| 邱晓芬, 汤冬娥, 虞海燕, 廖秋燕, 胡芷洋, 周俊, 赵鑫, 何慧燕, 梁灼健, 许承明, 杨明, 戴勇. 基于单细胞ATAC测序技术对18-三体综合征染色质开放性区域转录因子的分析. 遗传, 2021, 43(1): 74-83. | |
| [100] |
Ameen M, Sundaram L, Shen MC, Banerjee A, Kundu S, Nair S, Shcherbina A, Gu MX, Wilson KD, Varadarajan A, Vadgama N, Balsubramani A, Wu JC, Engreitz JM, Farh K, Karakikes I, Wang KC, Quertermous T, Greenleaf WJ, Kundaje A. Integrative single-cell analysis of cardiogenesis identifies developmental trajectories and non-coding mutations in congenital heart disease. Cell, 2022, 185(26): 4937-4953.e23.
doi: 10.1016/j.cell.2022.11.028 pmid: 36563664 |
| [101] |
Gerli MFM, Calà G, Beesley MA, Sina B, Tullie L, Sun KY, Panariello F, Michielin F, Davidson JR, Russo FM, Jones BC, Lee DDH, Savvidis S, Xenakis T, Simcock IC, Straatman-Iwanowska AA, Hirst RA, David AL, O'Callaghan C, Olivo A, Eaton S, Loukogeorgakis SP, Cacchiarelli D, Deprest J, Li VSW, Giobbe GG, De Coppi P. Single-cell guided prenatal derivation of primary fetal epithelial organoids from human amniotic and tracheal fluids. Nat Med, 2024, 30(3): 875-887.
doi: 10.1038/s41591-024-02807-z pmid: 38438734 |
| [102] |
Dixit A, Parnas O, Li BY, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, Adamson B, Norman TM, Lander ES, Weissman JS, Friedman N, Regev A. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell, 2016, 167(7): 1853-1866.e17.
doi: S0092-8674(16)31610-5 pmid: 27984732 |
| [103] | Santinha AJ, Klingler E, Kuhn M, Farouni R, Lagler S, Kalamakis G, Lischetti U, Jabaudon D, Platt RJ. Transcriptional linkage analysis with in vivo AAV-Perturb-seq. Nature, 2023, 622(7982): 367-375. |
| [104] |
Tran T, Song CJ, Nguyen T, Cheng SY, McMahon JA, Yang R, Guo QY, Der B, Lindström NO, Lin DC, McMahon AP. A scalable organoid model of human autosomal dominant polycystic kidney disease for disease mechanism and drug discovery. Cell Stem Cell, 2022, 29(7): 1083-1101.e7.
doi: 10.1016/j.stem.2022.06.005 pmid: 35803227 |
| [105] | Li C, Fleck JS, Martins-Costa C, Burkard TR, Themann J, Stuempflen M, Peer AM, Vertesy Á, Littleboy JB, Esk C, Elling U, Kasprian G, Corsini NS, Treutlein B, Knoblich JA. Single-cell brain organoid screening identifies developmental defects in autism. Nature, 2023, 621(7978): 373-380. |
| [106] | Bendixen L, Jensen TI, Bak RO. CRISPR-Cas-mediated transcriptional modulation: the therapeutic promises of CRISPRa and CRISPRi. Mol Ther, 2023, 31(7): 1920-1937. |
| [107] |
Nuñez JK, Chen J, Pommier GC, Cogan JZ, Replogle JM, Adriaens C, Ramadoss GN, Shi QM, Hung KL, Samelson AJ, Pogson AN, Kim JYS, Chung A, Leonetti MD, Chang HY, Kampmann M, Bernstein BE, Hovestadt V, Gilbert LA, Weissman JS. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell, 2021, 184(9): 2503-2519.e17.
doi: 10.1016/j.cell.2021.03.025 pmid: 33838111 |
| [108] | Jin X, Simmons SK, Guo A, Shetty AS, Ko M, Nguyen L, Jokhi V, Robinson E, Oyler P, Curry N, Deangeli G, Lodato S, Levin JZ, Regev A, Zhang F, Arlotta P. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science, 2020, 370(6520): eaaz6063. |
| [109] |
Chen XL, Yang WT, Roberts CWM, Zhang JH. Developmental origins shape the paediatric cancer genome. Nat Rev Cancer, 2024, 24(6): 382-398.
doi: 10.1038/s41568-024-00684-9 pmid: 38698126 |
| [110] | Ma XT, Liu Y, Liu YL, Alexandrov LB, Edmonson MN, Gawad C, Zhou X, Li YJ, Rusch MC, Easton J, Huether R, Gonzalez-Pena V, Wilkinson MR, Hermida LC, Davis S, Sioson E, Pounds S, Cao XY, Ries RE, Wang ZM, Chen X, Dong L, Diskin SJ, Smith MA, Guidry Auvil JM, Meltzer PS, Lau CC, Perlman EJ, Maris JM, Meshinchi S, Hunger SP, Gerhard DS, Zhang JH. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature, 2018, 555(7696): 371-376. |
| [111] | Luo ZL, Xia MY, Shi W, Zhao CT, Wang JJ, Xin DZ, Dong XR, Xiong Y, Zhang F, Berry K, Ogurek S, Liu XZ, Rao R, Xing R, Wu LMN, Cui SY, Xu LL, Lin YF, Ma WK, Tian SW, Xie Q, Zhang L, Xin M, Wang XT, Yue F, Zheng HZ, Liu YP, Stevenson CB, de Blank P, Perentesis JP, Gilbertson RJ, Li H, Ma J, Zhou WH, Taylor MD, Lu QR. Human fetal cerebellar cell atlas informs medulloblastoma origin and oncogenesis. Nature, 2022, 612(7941): 787-794. |
| [112] |
Kameneva P, Artemov AV, Kastriti ME, Faure L, Olsen TK, Otte J, Erickson A, Semsch B, Andersson ER, Ratz M, Frisén J, Tischler AS, de Krijger RR, Bouderlique T, Akkuratova N, Vorontsova M, Gusev O, Fried K, Sundström E, Mei SL, Kogner P, Baryawno N, Kharchenko PV, Adameyko I. Single-cell transcriptomics of human embryos identifies multiple sympathoblast lineages with potential implications for neuroblastoma origin. Nat Genet, 2021, 53(5): 694-706.
doi: 10.1038/s41588-021-00818-x pmid: 33833454 |
| [113] |
Bedoya-Reina OC, Li W, Arceo M, Plescher M, Bullova P, Pui H, Kaucka M, Kharchenko P, Martinsson T, Holmberg J, Adameyko I, Deng Q, Larsson C, Juhlin CC, Kogner P, and Schlisio S. Single-nuclei transcriptomes from human adrenal gland reveal distinct cellular identities of low and high-risk neuroblastoma tumors. Nat Commun, 2021, 12(1): 5309.
doi: 10.1038/s41467-021-24870-7 pmid: 34493726 |
| [114] | Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen YW, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, Lim DA. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science, 2017, 355(6320): aah7111. |
| [115] |
Kang L, Chen JY, Gao SR. Historical review of reprogramming and pluripotent stem cell research in China. Hereditas(Beijing), 2018, 40(10): 825-840.
doi: 10.16288/j.yczz.18-209 pmid: 30369467 |
| 康岚, 陈嘉瑜, 高绍荣. 中国细胞重编程和多能干细胞研究进展. 遗传, 2018, 40(10): 825-840. | |
| [116] |
Mills RJ, Parker BL, Quaife-Ryan GA, Voges HK, Needham EJ, Bornot A, Ding M, Andersson H, Polla M, Elliott DA, Drowley L, Clausen M, Plowright AT, Barrett IP, Wang QD, James DE, Porrello ER, Hudson JE. drug screening in human PSC-cardiac organoids identifies pro-proliferative compounds acting via the mevalonate pathway. Cell Stem Cell, 2019, 24(6): 895-907.e6.
doi: S1934-5909(19)30108-0 pmid: 30930147 |
| [117] |
Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui BX, Gold JD, Wu JC. Chemically defined generation of human cardiomyocytes. Nat Methods, 2014, 11(8): 855-60.
doi: 10.1038/nmeth.2999 pmid: 24930130 |
| [118] |
Saelens W, Cannoodt R, Todorov H, Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol, 2019, 37(5): 547-554.
doi: 10.1038/s41587-019-0071-9 pmid: 30936559 |
| [119] |
Van de Sande B, Flerin C, Davie K, De Waegeneer M, Hulselmans G, Aibar S, Seurinck R, Saelens W, Cannoodt R, Rouchon Q, Verbeiren T, De Maeyer D, Reumers J, Saeys Y, Aerts S. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc, 2020, 15(7): 2247-2276.
doi: 10.1038/s41596-020-0336-2 pmid: 32561888 |
| [120] |
Armingol E, Officer A, Harismendy O, Lewis NE. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet, 2021, 22(2): 71-88.
doi: 10.1038/s41576-020-00292-x pmid: 33168968 |
| [121] |
Shen S, Sun YLZ, Matsumoto M, Shim WJ, Sinniah E, Wilson SB, Werner T, Wu ZX, Bradford ST, Hudson J, Little MH, Powell J, Nguyen Q, Palpant NJ. Integrating single-cell genomics pipelines to discover mechanisms of stem cell differentiation. Trends Mol Med, 2021, 27(12): 1135-1158.
doi: 10.1016/j.molmed.2021.09.006 pmid: 34657800 |
| [122] |
Gonçalves CA, Larsen M, Jung S, Stratmann J, Nakamura A, Leuschner M, Hersemann L, Keshara R, Perlman S, Lundvall L, Thuesen LL, Hare KJ, Amit I, Jørgensen A, Kim YH, Del Sol A, Grapin-Botton A. A 3D system to model human pancreas development and its reference single-cell transcriptome atlas identify signaling pathways required for progenitor expansion. Nat Commun, 2021, 12(1): 3144.
doi: 10.1038/s41467-021-23295-6 pmid: 34035279 |
| [123] |
Haniffa M, Maartens A, Teichmann SA. How developmental cell atlases inform stem cell embryo models. Nat Methods, 2023, 20(12): 1849-1851.
doi: 10.1038/s41592-023-02072-x pmid: 38057509 |
| [124] |
Changmeng Z, Hongfei W, Cheung MC, Chan YS, Shea GK. Revealing the developmental origin and lineage predilection of neural progenitors within human bone marrow via single-cell analysis: implications for regenerative medicine. Genome Med, 2023, 15(1): 66.
doi: 10.1186/s13073-023-01224-0 pmid: 37667405 |
| [125] |
Mikryukov AA, Mazine A, Wei B, Yang DH, Miao YF, Gu MX, Keller GM. BMP10 signaling promotes the development of endocardial cells from human pluripotent stem cell-derived cardiovascular progenitors. Cell Stem Cell, 2021, 28(1): 96-111.e7.
doi: 10.1016/j.stem.2020.10.003 pmid: 33142114 |
| [126] |
Raff RA. Written in stone: fossils, genes and evo-devo. Nat Rev Genet, 2007, 8(12): 911-20.
pmid: 18007648 |
| [127] | Müller GB. Evo-devo: extending the evolutionary synthesis. Nat Rev Genet, 2007, 8(12): 943-9. |
| [128] | Sepp M, Leiss K, Murat F, Okonechnikov K, Joshi P, Leushkin E, Spänig L, Mbengue N, Schneider C, Schmidt J, Trost N, Schauer M, Khaitovich P, Lisgo S, Palkovits M, Giere P, Kutscher LM, Anders S, Cardoso- Moreira M, Sarropoulos I, Pfister SM, Kaessmann H. Cellular development and evolution of the mammalian cerebellum. Nature, 2024, 625(7996): 788-796. |
| [129] |
Zhong JX, Aires R, Tsissios G, Skoufa E, Brandt K, Sandoval-Guzmán T, Aztekin C. Multi-species atlas resolves an axolotl limb development and regeneration paradox. Nat Commun, 2023, 14(1): 6346.
doi: 10.1038/s41467-023-41944-w pmid: 37816738 |
| [130] |
Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol, 1983, 100(1): 64-119.
doi: 10.1016/0012-1606(83)90201-4 pmid: 6684600 |
| [131] |
Blain R, Couly G, Shotar E, Blévinal J, Toupin M, Favre A, Abjaghou A, Inoue M, Hernández-Garzón E, Clarençon F, Chalmel F, Mazaud-Guittot S, Giacobini P, Gitton Y, Chedotal A. A tridimensional atlas of the developing human head. Cell, 2023, 186(26): 5910-5924.e17.
doi: 10.1016/j.cell.2023.11.013 pmid: 38070509 |
| [132] | de Bakker BS, de Jong KH, Hagoort J, de Bree K, Besselink CT, Veldhuis T, Bais B, Schildmeijer R, Ruijter JM, Oostra RJ, Christoffels VM, Moorman AF. An interactive three-dimensional digital atlas and quantitative database of human development. Science, 2016, 354(6315): aag0053. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
