Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (3): 278-286.doi: 10.16288/j.yczz.20-015
• Review • Previous Articles Next Articles
Molecular genetic mechanisms of interaction between host plants and pathogens
Dewei Yang1,3, Shengping Li2,3, Haitao Cui2,3, Shenghao Zou3, Wei Wang2,3( )
)
- 1. Institute of Rice, Fujian Academy of Agricultural Sciences, Fuzhou 350018, China
2. College of Agriculture, Fujian Agriculture and Forestry University, Fuzhou 350002, China
3. Plant Immunity Center, Fujian Agriculture and Forestry University, Fuzhou 350002, China
-
Received:2020-01-13Revised:2020-02-19Online:2020-03-20Published:2020-03-01 -
Contact:Wang Wei E-mail:vic_0214@163.com -
Supported by:Supported by the Youth Technology Innovation Team of the Fujian Academy of Agricultural Sciences No(STIT2017-3-3);the Special Fund for Agro-scientific Research in the Public Interest of Fujian Province No(2017R1021-2);Fujian Provincial Natural Science Foundation No(2019J01102);the Free Exploration Project of the Fujian Academy of Agricultural Sciences No(AA2018-21)
Cite this article
Dewei Yang, Shengping Li, Haitao Cui, Shenghao Zou, Wei Wang. Molecular genetic mechanisms of interaction between host plants and pathogens[J]. Hereditas(Beijing), 2020, 42(3): 278-286.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Supplementary Table 1
Plant disease resistance genes and avirulence genes"
| 无毒基因 | 病原菌 | 寄主 | 抗性基因 | 参考文献链接 |
|---|---|---|---|---|
| P3 | Soybean mosaic virus | Glycine max | 3gG2 | |
| Avr9 | Cladosporium fulvum (Syn. Passalora fulva) | Solanum lycopersicum | 9DC1 | |
| Avr9 | Cladosporium fulvum (Syn. Passalora fulva) | Solanum lycopersicum | 9DC2(Syn. 9DC) | |
| Avr9 | Cladosporium fulvum (Syn. Passalora fulva) | Solanum lycopersicum | 9DC3 | |
| Sphinganine-analog mycotoxins | Alternaria alternata | Solanum lycopersicum | Asc-1 | |
| Not identification | Pseudoperonospora cubensis | Cucumis melo L. | At1 | |
| Not identification | Pseudoperonospora cubensis | Cucumis melo L. | At2 | |
| Not identification | Clover yellow vein virus | Phaseolus vulgaris | bc-3 | |
| Not identification | Turnip mosaic virus (TuMV) | Brasica pekinensis Rupr | BcTuR3 | |
| Not identification | Nilaparvata lugens | Oryza sativa L. | Bph1/9-1 (Syn. Bph1/ Bph10/Bph18/Bph21) | |
| Not identification | Nilaparvata lugens | Oryza sativa L. | Bph1/9-2 (Syn. Bph2/Bph26) | |
| Not identification | Nilaparvata lugens | Oryza sativa L. | Bph1/9-7 (Syn. Bph7;Syn. Bph9) | |
| Not identification | Nilaparvata lugens | Oryza sativa L. | Bph14 | |
| AvrBs2 | Xanthomonas axonopodis | Solanum lycopersicum | Bs2 | |
| AvrHah1 | Xanthomonas gardneri | Piper nigrum L. | Bs3 | |
| Hax4 | Xanthomonas campestris | Solanum lycopersicum | Bs4 | |
| AvrBs4 | Xanthomonas campestris | Piper nigrum L. | Bs4C-R | |
| Putative PGN | Pseudomonas syringae | Solanum lycopersicum | Bti9 | |
| Not identification | Ralstonia solanacearum | Nicotiana L. | CaLRR51 | |
| Not identification | Meloidogyne incognita | Piper nigrum L. | CaMi | |
| Not identification | Phakopsora pachyrhizi | Glycine?max | CcRpp1 | |
| Avr4 | Cladosporium fulvum (Syn. Passalora fulva) | Solanum lycopersicum | Cf-4 (Syn. Hcr9-4D) | |
| Avr5 | Cladosporium fulvum (Syn. Passalora fulva) | Solanum lycopersicum | Cf-5 (Syn. Hcr2-5C) | |
| Avr9 | Cladosporium fulvum (Syn. Passalora fulva) | Solanum lycopersicum | Cf-9 (Syn. Hcr9-9C) | |
| Not identification | - | Nicotiana benthamiana | Cf-9B (Hcr9-9B) | |
| Avr9B | Cladosporium fulvum (Syn. Passalora fulva) | Solanum lycopersicum | Cf-9B (Syn. Hcr9-9B) | |
| Not identification | Heterodera avenae | Triticum aestivum | Cre1 | |
| Not identification | Heterodera avenae | Triticum aestivum | Cre3 | |
| Not identification | Clover yellow vein virus | Arabidopsis thaliana | cum1-1 | |
| Not identification | Cuscuta reflexa | Solanum lycopersicum | CuRe1 | |
| Coat protein | Mungbean Yellow Mosaic India Virus (MYMIV) | Vigna mungo | CYR1 | |
| Not identification | - | Arabidopsis thaliana | Dm1 (Uk-3) | |
| Not identification | Potato virus Y (PVY) | Solanum tuberosum L. | Eva1 | |
| Not identification | Plantago Asiatica Mosaic Virus (PIAMV) | Arabidopsis thaliana | EXA1 | |
| AvrFom1 | Fusarium oxysporum | Cucumis melo L. | Fom-1 (Syn. RGH9) | |
| AvrFom2 | Fusarium oxysporum | Cucumis melo L. | Fom-2 | |
| Not identification | Verticillium dahliae | Gossypium spp | GbVE | |
| Not identification | Xanthomonas oryzae | Oryza sativa L. | GH3-2 | |
| Not identification | Xanthomonas oryzae | Oryza sativa L. | GH3-8 | |
| Not identification | Heterodera glycines | Glycine max | GmSNAP18 (Syn. Rhg1-a) | |
| RBP-1 | Globodera pallida | Solanum tuberosum L. | Gpa2 | |
| Not identification | Globodera rostochiensis | Solanum tuberosum L. | Gro1-4 | |
| Not identification | Cercospora zeae-maydis | Zea mays | GST23 | |
| Not identification | Plasmopara halstedii | Sunflower | Ha-NTIR11g | |
| Avr4E | Cladosporium fulvum (Syn. Passalora fulva) | Solanum lycopersicum | Hcr9-4E | |
| Not identification | - | Nicotiana benthamiana | Hcr9-9A | |
| Not identification | Cladosporium fulvum (Syn. Passalora fulva) | Solanum lycopersicum | Hcr9-9E | |
| Not identification | Venturia inaequalis | Apple | HcrVf2 | |
| Not identification | Globodera rostochiensis | Solanum lycopersicum | Hero A | |
| Ave1 | Verticillium dahliae | Arabidopsis thaliana | HLVe1-2A | |
| HC toxin | Cochliobolus carbonum | Zea mays | Hm1 | |
| Coat protein | Turnip crinckle virus | Arabidopsis thaliana | Hrt1 | |
| Not identification | Heterodera schachtii | Beta vulgaris L. | Hs1(pro-1) | |
| Not identification | Pseudomonas syringae | Nicotiana L. | Hth | |
| Not identification | Exserohilum turcicum | Zea mays | Htn1 (Syn. ZmWAK-RLK1) | |
| HC toxin | Cochliobolus carbonum | Hordeum?vulgare | HvHm1 | |
| Avr1 (Syn. Six4) | Fusarium oxysporum | Solanum lycopersicum | I | |
| Avr2 (Six3) | Fusarium oxysporum | Solanum lycopersicum | I2 | |
| Not identification | Fusarium oxysporum | Solanum lycopersicum | I-7 | |
| Not identification | Diabrotica virgifera | Zea mays | IPD072Aa | |
| Not identification | Botrytis cinerea | Solanum lycopersicum | IVR | |
| Not identification | Tobacco Etch Virus | Arabidopsis thaliana | JAX1 | |
| Coat protein | Tobamovirus | Piper nigrum L. | L1a | |
| Not identification | Melampsora lini | Linum usitatissimum | L2 | |
| Coat protein | Tobamovirus | Piper nigrum L. | L2b | |
| Not identification | Melampsora lini | Linum usitatissimum | L3 | |
| AvrL567-E | Melampsora lini | Linum usitatissimum | L5 | |
| Not identification | Melampsora lini | Linum usitatissimum | L8 | |
| Not identification | Striga | Sorghum | LGS1 | |
| Victorin | Cochliobolus victoriae | Arabidopsis thaliana | LOV1 | |
| AVR1 | Puccinia triticina | Triticum aestivum | Lr1 | |
| AvrLr21 | Puccinia triticina | Triticum aestivum | Lr21 | |
| AVR1 | Puccinia triticina | Triticum aestivum | Lr22a | |
| Not identification | Puccinia striiformis | Triticum aestivum | Lr34 (syn. Yr18/Pm38/Ltn1) | |
| Not identification | Puccinia striiformis | Triticum aestivum | Lr67 (syn. Pm46/ Sr55/Yr46/Ltn3) | |
| Not identification | Turnip mosaic virus (TuMV) | Arabidopsis thaliana | lsp1-1 | |
| AvrM | Melampsora lini | Linum usitatissimum | M | |
| Not identification | Meloidogyne | Prunus cerasifera Ehrh | Ma | |
| Not identification | Meloidogyne | Solanum lycopersicum | Mi-1 | |
| AVRa1 | Blumeria graminis | Hordeum?vulgare | Mla1 | |
| AVRa10 | Blumeria graminis | Hordeum?vulgare | Mla10 | |
| AvrMla12 | Blumeria graminis | Hordeum?vulgare | Mla12 | |
| AVRa13 | Blumeria graminis | Hordeum?vulgare | Mla13 | |
| AvrMla6 | Blumeria graminis | Hordeum?vulgare | Mla6 | |
| AvrMla7 | Blumeria graminis | Hordeum?vulgare | Mla7 | |
| Not identification | Blumeria graminis | Hordeum?vulgare | Mla8 | |
| Not identification | Blumeria graminis | Hordeum?vulgare | Mla9 | |
| Not identification | Lettuce Mosaic Virus | Lactuca sativa L. var. ramosa Hort | Mo-1 | |
| Not identification | Diplocarpon rosae | Rosa rugosa Thunb | muRdr1A (Syn. Rdr1) | |
| AvrPto | Pseudomonas syringae | Nicotiana benthamiana | NbPrf (Niben101Scf00650g02002XLOC) | |
| FoAve1 | Fusarium oxysporum | Nicotiana glutinosa | NgVe1 | |
| Not identification | Xanthomonas oryzae | Oryza sativa L. | OsAPX8 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | OsCul3a | |
| AvrP | Melampsora lini | Linum usitatissimum | P | |
| AvrP123 | Melampsora lini | Linum usitatissimum | P2 | |
| Not identification | Heterobasidion parviporum | Picea abies | PaLAR3 | |
| Not identification | Pantoea stewartii | Zea mays | pan1 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pb1 | |
| AvrBs3Δ16 | Xanthomonas campestris | Piper nigrum L. | pBs3-E | |
| Pc-toxin | Periconia circinata | Sorghum bicolor (L.) Moench | Pc | |
| Not identification | Tomato yellow leaf curl virus | Solanum lycopersicum | Pelo (Syn. Ty-5) | |
| Not identification | Phytophthora infestans | Solanum lycopersicum | Ph-3 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi1-5 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi2 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | pi21 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi25 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi35 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi36 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi37 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi50 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi5-2 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi54 (Syn. Pik-k(h)) | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi55(t) | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi64 | |
| AvrPi9 | Magnaporthe oryzae | Oryza sativa L. | Pi9 | |
| AvrPiB | Magnaporthe oryzae | Oryza sativa L. | Pib | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pi-d2 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pid3 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pid3-A4 | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | PigmR (Syn. PigmR6) | |
| AvrPii | Magnaporthe oryzae | Oryza sativa L. | Pii | |
| Avr-PikE | Magnaporthe oryzae | Oryza sativa L. | Pik-1 | |
| Avr-PikA | Magnaporthe oryzae | Oryza sativa L. | Pik-h | |
| Avr-PikD | Magnaporthe oryzae | Oryza sativa L. | Pik-p1 | |
| Avr-PikD | Magnaporthe oryzae | Oryza sativa L. | Pik-s | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pish | |
| Not identification | Magnaporthe oryzae | Oryza sativa L. | Pit | |
| AvrPita2 | Magnaporthe oryzae | Oryza sativa L. | Pi-ta | |
| AvrPiz-t | Magnaporthe oryzae | Oryza sativa L. | Piz-t | |
| BgsE-5845 | Blumeria graminis | Triticum aestivum | Pm2 | |
| AvrPm3a2/f2 | Blumeria graminis | Triticum aestivum | Pm3a | |
| AvrPm3b | Blumeria graminis | Triticum aestivum | Pm3b | |
| Not identification | Blumeria graminis | Triticum aestivum | Pm3c | |
| Not identification | Blumeria graminis | Triticum aestivum | Pm3d | |
| Not identification | Blumeria graminis | Triticum aestivum | Pm3e | |
| AvrPm3a2/f2 | Blumeria graminis | Triticum aestivum | Pm3f | |
| Not identification | Blumeria graminis | Triticum aestivum | Pm3g | |
| Not identification | Blumeria graminis | Triticum aestivum | Pm3k | |
| Not identification | Potato virus Y (PVY) | Solanum lycopersicum | Pot-1 | |
| avrPtoB, AvrPto (B728a) | Pseudomonas syringae | Solanum lycopersicum | Prf | |
| avrBs3 | Xanthomonas citri | Citrus X paradise | ProBs314EBE::avrGf1 | |
| Not identification | Papaya ring-spot virus | Cucumis melo L. | Prv1 (Syn. RGH10) | |
| 2a | Cucumber mosaic virus (CMV) | Phaseolus vulgaris | PvCMR1 (Syn. RT4-4) | |
| Not identification | Potato virus Y (PVY) | Piper nigrum L. | Pvr21 | |
| RNA-dependent RNA polymerase (NIb) | Potato virus Y (PVY) | Nicotiana benthamiana | Pvr4 | |
| Not identification | Pepper Veinal Mottle Virus | Piper nigrum L. | pvr6 | |
| BV1 | Bean Dwarf Mosaic Virus | Phaseolus vulgaris | PvVTT1 | |
| Not identification | Magnaporthe grisea | Zea mays | qMdr9.02 (Syn. ZmCCoAOMT2) | |
| Not identification | Fusarium graminearum | Zea mays | qRfg1 (Syn. ZmCCT) | |
| Avr1 | Phytophthora infestans | Solanum tuberosum L. | R1 | |
| Avr2 | Phytophthora infestans | Solanum tuberosum L. | R2 | |
| Avr3a KI | Phytophthora infestans | Solanum tuberosum L. | R3a | |
| Avr3b | Phytophthora infestans | Solanum tuberosum L. | R3b | |
| Avr8 | Phytophthora infestans | Solanum tuberosum L. | R8 (Syn. Rpi-smira2) | |
| Not identification | Albugo candida | Arabidopsis thaliana | RAC1 | |
| Avrblb1 (Syn. IPI-O/PexRD) | Phytophthora infestans | Solanum tuberosum L. | Rb (Syn. Rpi-blb1) | |
| HopBA1 | Pseudomonas syringae | Arabidopsis thaliana | RBA1 | |
| Not identification | Glomerella graminicola (Syn. Colletotrichum graminicola) | Zea mays | Rcg1 | |
| Not identification | Colletotrichum trifolii | Lotuscorniculatus L. | RCT1 | |
| Coat protein | Cucumber mosaic virus (CMV) | Arabidopsis thaliana | Rcy1 | |
| Not identification | Pyrenophora graminea | Hordeum vulgare | Rdg2a | |
| Not identification | Turnip mosaic virus (TuMV) | Brassica oleracea L. var.capitata L. | retr01 | |
| Not identification | Fusarium oxysporum | Arabidopsis thaliana | RFO1 | |
| Not identification | Fusarium oxysporum | Arabidopsis thaliana | RFO3 | |
| AvrLr10 | Puccinia triticina | Triticum aestivum | Rga2 | |
| Not identification | - | Triticum aestivum | RGA2a | |
| Avr-Pia | Magnaporthe oryzae | Oryza sativa L. | RGA5-A (Syn. Pia-1) | |
| Avr3 | Bremia lactucae | Lactuca sativa L. var. ramosa Hort | RGC2B (Syn. Dm3) | |
| Not identification | Heterodera glycines | Glycine max | Rhg1-b | |
| Not identification | Leptosphaeria maculans | Arabidopsis thaliana | RLM1A | |
| AvrLm2 | Leptosphaeria maculans | Brassica campestris L. | RLM2 | |
| Not identification | Alternaria brassicae | Arabidopsis thaliana | RLM3 | |
| XopQ | Xanthomonas campestris | Nicotiana benthamiana | Roq1 | |
| Complex of PGTG_10537.2 and PGTG_16791 | Puccinia graminis | Hordeum vulgare | Rpg1 | |
| AvrB | Pseudomonas savastanoi | Glycine max | Rpg1-b | |
| AvrRpm1 | Pseudomonas syringae | Glycine max | Rpg1r | |
| Not identification | Puccinia graminis | Hordeum vulgare | Rpg5 | |
| AvrLr10 | Puccinia sorghi | Zea mays | Rpi1-D | |
| Avr2 | Phytophthora infestans | Solanum tuberosum L. | Rpi-abpt | |
| Not identification | Phytophthora infestans | Solanum tuberosum L. | Rpi-amr3i | |
| Avrblb2 (Syn. PexRD39/PexRD40) | Phytophthora infestans | Solanum tuberosum L. | Rpi-blb2 | |
| Avr2 | Phytophthora infestans | Solanum tuberosum L. | Rpi-blb3 | |
| Avrblb1 (Syn. IPI-O/PexRD) | Phytophthora infestans | Solanum tuberosum L. | Rpi-pta1 | |
| Avr2 | Phytophthora infestans | Solanum tuberosum L. | Rpi-snk1.1 | |
| Avrblb1 (Syn. IPI-O/PexRD) | Phytophthora infestans | Solanum tuberosum L. | Rpi-sto1 | |
| Avrvnt1 | Phytophthora infestans | Solanum tuberosum L. | Rpi-vnt1.1 | |
| AvrRpm1, AvrB | Pseudomonas syringae | Arabidopsis thaliana | RPM1 | |
| ATR13-Emco5 | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | RPP13-Nd-1 | |
| Not identification | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | RPP13-Rld-2 | |
| ATR13-Maks9 | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | RPP13-UKID34 | |
| ATR13-Emco5 | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | RPP13-UKID37 | |
| Atr1Ndwsb | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | RPP1-EstA | |
| Atr1Ndwsb | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | Rpp1Nda | |
| Not identification | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | Rpp1-Wsa | |
| Atr1Ndwsb | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | Rpp1-Wsb | |
| Not identification | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | Rpp1-Wsc | |
| Atr1Ndwsb | Hyaloperonospora arabidopsidis | Arabidopsis thaliana | RPP1-ZdrA | |
| Not identification | Phakopsora pachyrhizi | Glycine max | Rpp4C4 | |
| AvrRpt2 | Phytophthora sojae | Glycine max | Rps1k-1 and/or Rps1k-2 | |
| AvrRpt2 | Pseudomonas syringae | Arabidopsis thaliana | RPS2 | |
| AvrPphB | Pseudomonas syringae | Arabidopsis thaliana | RPS5 | |
| HopA1 (hrmA/hopPsyA) | Pseudomonas syringae | Arabidopsis thaliana | RPS6 | |
| Not identification | Erysiphe cichoracearum (Syn. Golovinomyces cichoracearum) | Arabidopsis thaliana | Rpw8.1 | |
| Not identification | Erysiphe cichoracearum (Syn. Golovinomyces cichoracearum) | Arabidopsis thaliana | Rpw8.2 | |
| AvrRps4 (Syn. AvrPpiE) | Pseudomonas syringae | Arabidopsis thaliana | RRS1B | |
| AvrRps4 (Syn. AvrPpiE) | Pseudomonas syringae | Arabidopsis thaliana | RRS1-S | |
| Not identification | Xanthomonas campestris | Arabidopsis thaliana | RRS1ws-0 | |
| Not identification | Striga gesnerioides | Pisum sativum L. | RSG3-301 | |
| Not identification | Tobacco Etch Virus | Arabidopsis thaliana | Rtm3 | |
| Not identification | Golovinomyces cichoracearum | Arabidopsis thaliana | RTP1 | |
| Not identification | Watermelon Mosaic Virus | Arabidopsis thaliana | rwm1 | |
| Coat protein | Potato virus X (PVX) | Solanum tuberosum L. | Rx1 | |
| AvrRxo1-ORF1 | Xanthomonas oryzae | Zea mays | Rxo1 | |
| Not identification | Barley yellow mosaic virus | Hordeum vulgare | Rym-4 | |
| Not identification | Beet Necrotic Yellow Vein Virus (BNYVV) | Beta vulgaris L. | Rz2 | |
| Not identification | Bean yellow mosaic virus | Pisum sativum L. | sbm1 (Syn. wlv/cyv2) | |
| Not identification | Heterodera glycines | Glycine max | SHMT (Syn. Rhg4) | |
| Not identification | Pepper mottle virus | Solanum lycopersicum | SleIF4E1-G1485A | |
| Putative PGN | Pseudomonas syringae | Solanum lycopersicum | SlLyk13 | |
| SnTox1 | Stagonospora nodorum | Triticum aestivum | Snn1 (Syn. TaWAK1) | |
| Not identification | Puccinia graminis | Triticum aestivum | Sr22 | |
| Not identification | Puccinia graminis | Triticum aestivum | Sr33 (Syn. Rga1e) | |
| Not identification | Puccinia graminis | Triticum aestivum | Sr35 | |
| Not identification | Puccinia graminis | Triticum aestivum | Sr45 | |
| Not identification | Puccinia graminis | Triticum aestivum | Sr50 (Syn. ScRGA1-A/SrR) | |
| FoAve1 | Fusarium oxysporum | Solanum melongena L. | StoVe1 | |
| Not identification | Blumeria graminis | Triticum aestivum | Stpk-V (Pm21) | |
| CbAve1 | Cercospora beticola | Solanum tuberosum L. | StuVe1 | |
| Not identification | Rice Stripe Virus | Oryza sativa L. | STV11-R | |
| HopAI1 | Pseudomonas syringae | Arabidopsis thaliana | SUMM2 | |
| Nsm | Tomato spotted wilt virus | Solanum lycopersicum | Sw-5b | |
| Not identification | Blumeria graminis | Triticum aestivum | Taedr1 | |
| AvrB | Pseudomonas savastanoi | Arabidopsis thaliana | Tao1 | |
| Not identification | Sclerotinia sclerotiorum | Solanum lycopersicum | TcBI-1 | |
| RNA-dependent RNA polymerase | Tomato Mosaic Virus (ToMV) | Solanum lycopersicum | Tm-1 | |
| Movement protein | Tobacco mosaic virus (TMV) | Solanum lycopersicum | Tm-2 | |
| Not identification | Bemisia tabaci | Gossypium spp. | Tma12 | |
| Not identification | Blumeria graminis | Triticum aestivum | TmMLA1 | |
| Not identification | Pseudomonas syringae | Arabidopsis thaliana | TN13 | |
| Not identification | - | Arabidopsis thaliana | TN2,TN1,SOC3 | |
| ToxA | Stagonospora nodorum | Triticum aestivum | Tsn1 | |
| NSs | Tomato spotted wilt virus | Nicotiana benthamiana | Tsw | |
| Not identification | Tomato yellow leaf curl virus | Solanum lycopersicum | Ty-1 | |
| Not identification | Aphis gossypii | Cucumis melo L. | VAT | |
| CbAve1 | Cercospora beticola | Solanum lycopersicum | Ve1 | |
| Not identification | Callosobruchus chinensis | Leguminosae spp. | VrPGIP2 | |
| Not identification | Puccinia striiformis | Triticum aestivum | WCBP1 (Syn. YrL693) | |
| Not identification | Puccinia striiformis | Triticum aestivum | WKS1 (syn. Yr36 ) | |
| Tal3a mutant (containing full C-terminus) | Xanthomonas oryzae | Oryza sativa L. | Xa1 | |
| AvrXa10 | Xanthomonas oryzae | Oryza sativa L. | Xa10 | |
| AvrXa10 | Xanthomonas oryzae | Oryza sativa L. | Xa10E5 | |
| PthXo1 | Xanthomonas oryzae | Oryza sativa L. | xa13 | |
| RaxX; RaxX21-sY | Xanthomonas oryzae | Oryza sativa L. | XA21 | |
| AvrXa23 | Xanthomonas oryzae | Oryza sativa L. | Xa23 | |
| Not identification | Xanthomonas oryzae | Oryza sativa L. | xa25 | |
| AvrXa27 | Xanthomonas oryzae | Oryza sativa L. | Xa27 | |
| Not identification | Xanthomonas oryzae | Oryza sativa L. | Xa4 | |
| Not identification | Xanthomonas oryzae | Oryza sativa L. | xa5 | |
| Not identification | Potato virus Y (PVY) | Solanum tuberosum L. | Y-1 | |
| Not identification | Puccinia striiformis | Triticum aestivum | Yr10 | |
| HopZ1a (Syn. HopPsyH) | Pseudomonas syringae | Arabidopsis thaliana | ZAR1 | |
| Not identification | Sugarcane Mosaic Virus | Zea mays | ZmTrxh (Syn. Scmv1) | |
| Not identification | Sphacelotheca reiliana | Zea mays | ZmWAK1 (Syn. qHSR1) |
| [1] | Ausubel FM . Are innate immune signaling pathways in plants and animals conserved? Nat Immunol, 2005,6(10):973-979. |
| [2] | Jones JD, Dangl JL . The plant immune system. Nature, 2006,444(7117):323-329. |
| [3] | Dangl JL, Jones JD . Plant pathogens and integrated defence responses to infection. Nature, 2001,411(6839):826-833. |
| [4] | Gómez-Gómez L, Boller T . FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell, 2000, ( 6):1003-1011. |
| [5] | Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T . A flagellin- induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature, 2007,448(7152):497-500. |
| [6] | Sun YD, Li L, Macho AP, Han ZF, Hu ZH, Zipfel C, Zhou JM, Chai JJ . Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science, 2013,342(6158):624-628. |
| [7] | Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G . Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium- mediated transformation. Cell, 2006,125(4):749-760. |
| [8] | Boller T, Felix G . A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol, 2009,60:379-406. |
| [9] | Pfeilmeier S, George J, Morel A, Roy S, Smoker M, Stransfeld L, Downie JA, Peeters N, Malone JG, Zipfel C . Expression of the Arabidopsis thaliana immune receptor EFR in Medicago truncatula reduces infection by a root pathogenic bacterium, but not nitrogen-fixing rhizobial symbiosis. Plant Biotechnol J, 2019,17(3):569-579. |
| [10] | Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii- Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, Shibuya N . Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J, 2010,64(2):204-214. |
| [11] | Liu J, Liu B, Chen SF, Gong BQ, Chen LJ, Zhou Q, Xiong F, Wang ML, Feng DR, Li JF, Wang HB, Wang JF . A tyrosine phosphorylation cycle regulates fungal activation of a plant receptor Ser/Thr Kinase. Cell Host Microbe, 2018,23(2):241-253. |
| [12] | Ao Y, Li ZQ, Feng DR, Xiong F, Liu J, Li JF, Wang ML, Wang JF, Liu B, Wang HB . OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J, 2014,80(6):1072-1084. |
| [13] | Dixon MS, Jones DA, Keddie JS, Thomas CM, Harrison K, Jones JD . The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell, 1996,84(3):451-459. |
| [14] | Luderer R , Takken FLW, de Wit PJGM, Joosten MHA[J]. Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol Microbiol, 2002,45(3):875-884. |
| [15] | Dixon MS, Golstein C, Thomas CM, van Der Biezen EA, Jones JD . Genetic complexity of pathogen perception by plants: the example of Rcr3, a tomato gene required specifically by Cf-2. Proc Natl Acad Sci USA, 2000,97(16):8807-8814. |
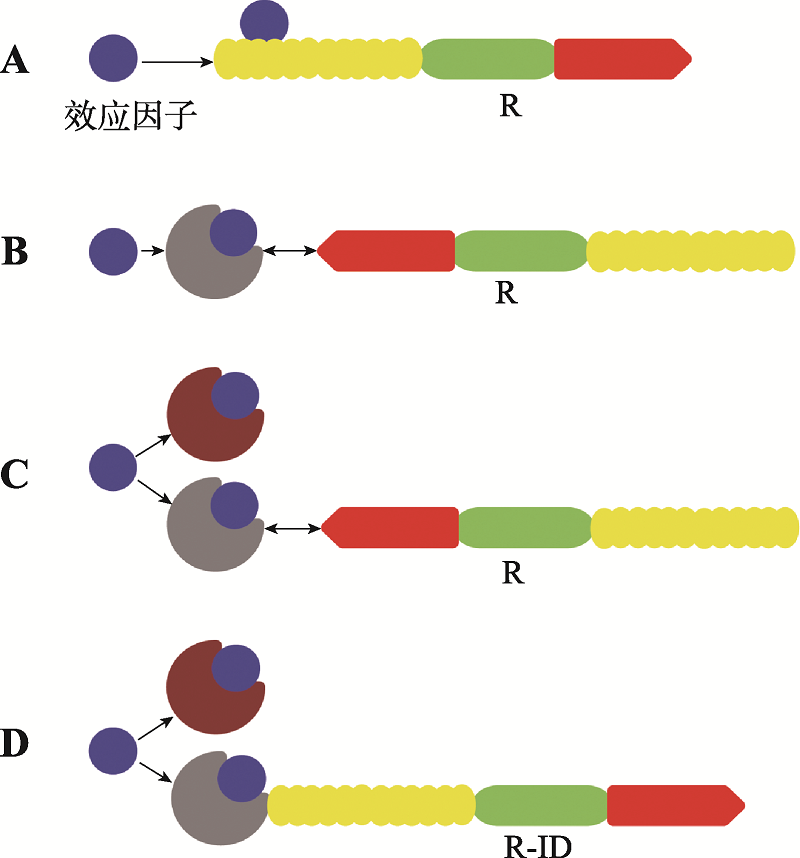
| [16] | van der Hoorn RAL, Kamoun S . From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell, 2008,20(8):2009-2017. |
| [17] | Johal GS, Briggs SP . Reductase activity encoded by the HM1 disease resistance gene in maize. Science, 1992,258(5084):985-987. |
| [18] | Kourelis J, van der Hoorn RAL . Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell, 2018,30(2):285-299. |
| [19] | Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ . Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc Natl Acad Sci USA, 2005,102(6):2087-2092. |
| [20] | Axtell MJ, Staskawicz BJ . Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell, 2003,112(3):369-377. |
| [21] | Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y . Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA, 2003,100(13):8024-8029. |
| [22] | Keen NT . Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet, 1990,24:447-463. |
| [23] | Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J, 2000,19(15):4004-4014. |
| [24] | Saur IM, Bauer S, Kracher B, Lu X, Franzeskakis L, Müller MC, Sabelleck B, Kümmel F, Panstruga R, Maekawa T, Schulze-Lefert P . Multiple pairs of allelic MLA immune receptor-powdery mildew AVRA effectors argue for a direct recognition mechanism. eLife, 2019,e44471. |
| [25] | van der Biezen EA, Jones JD . Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci, 1998,23(12):454-456. |
| [26] | Tasset C, Bernoux M, Jauneau A, Pouzet C, Brière C, Kieffer-Jacquinod S, Rivas S, Marco Y, Deslandes L . Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R- mediated immunity in Arabidopsis. PLoS Pathog, 2010,6(11):1-14. |
| [27] | Mackey D, Holt BF, Wiig A, Dangl JL . RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell, 2002,108(6):743-754. |
| [28] | Day B, Dahlbeck D, Huang J, Chisholm ST, Li D, Staskawicz BJ . Molecular basis for the RIN4 negative regulation of RPS2 disease resistance. Plant Cell, 2005,17(4):1292-1305. |
| [29] | Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G . The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell, 2006,18(2):465-476. |
| [30] | Dodds PN, Rathjen JP . Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet, 2010,11(8):539-548. |
| [31] | Zhang J, Li W, Xiang TT, Liu ZX, Laluk K, Ding XJ, Zou Y, Gao MH, Zhang XJ, Chen S, Mengiste T, Zhang YL, Zhou JM . Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe, 2010,7(4):290-301 |
| [32] | Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW . Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science, 2003,301(5637):1230-1233. |
| [33] | Césari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, Morel JB, Fournier E, Tharreau D, Terauchi R, Kroj T . The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell, 2013,25(4):1463-1481. |
| [34] | Maqbool A, Saitoh H, Franceschetti M, Stevenson CEM, Uemura A, Kanzaki H, Kamoun S, Terauchi R, Banfield MJ . Structural basis of pathogen recognition by an integrated HMA domain in a plant NLR immune receptor. eLife, 2015,4:e08709. |
| [35] | Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M . Loss of function of a proline-containing protein confers durable disease resistance in rice. Science, 2009,325(5943):998-1001. |
| [36] | Le Roux C, Huet G, Jauneau A, Camborde L, Trémousaygue D, Kraut A, Zhou B, Levaillant M, Adachi H, Yoshioka H, Raffaele S, Berthomé R, Couté Y, Parker JE, Deslandes L . A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell, 2015,161(5):1074-1088. |
| [37] | Ma ZC, Zhu L, Song TQ, Wang Y, Zhang Q, Xia YQ, Qiu M, Lin YC, Li HY, Kong L, Fang YF, Ye WW, Wang Y, Dong SM, Zheng XB, Tyler BM, Wang YC . A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science, 2017,355(6326):710-714. |
| [38] | Schornack S, Meyer A, Römer P, Jordan T, Lahaye T . Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J Plant Physiol, 2006,163(3):256-272. |
| [39] | Gu KY, Yang B, Tian DS, Wu LF, Wang DJ, Sreekala C, Yang F, Chu ZQ, Wang GL, White FF, Yin ZC . R gene expression induced by a type-III effector triggers disease resistance in rice. Nature, 2005,435(7045):1122-1125. |
| [40] | Römer P, Hahn S, Jordan T, Strauss T, Bonas U, Lahaye T . Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science, 2007,318(5850):645-648. |
| [41] | Chen H, Chen J, Li M, Chang M, Xu KM, Shang ZH, Zhao Y, Palmer I, Zhang YQ, McGill J, Alfano JR, Nishimura MT, Liu FQ, Fu ZQ. A bacterial type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe, 2017,22(6):777-788. |
| [42] | Lopez VA, Park BC, Nowak D, Sreelatha A, Zembek P, Fernandez J, Servage KA, Gradowski M, Hennig J, Tomchick DR, Pawłowski K, Krzymowska M, Tagliabracci VS. A bacterial effector mimics a host HSP90 client to undermine immunity. 2019, Cell, 179(1): 205- 218.e21. |
| [43] | Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U . Breaking the code of DNA binding specificity of TAL-type III effectors. Science, 2009; 326(5959):1509-1512. |
| [44] | He F, Wang CC, Wang FQ, Yang L . Interaction mechanism between plant resistance gene and pathogen avirulence gene. Chin J Cell Biol, 2011,33(9):1037-1044. |
| 何锋, 王长春, 王锋青, 杨玲 . 植物抗病基因(R)与病原物无毒基因(Avr)相互作用机制的研究进展. 中国细胞生物学学报, 2011,33(9):1037-1044. | |
| [45] | Yang DW, Wang M, Hang LB, Tang DZ, Li SP . Progress of cloning and breeding application of blast resistance genes in rice and avirulence genes in blast fungi. Chin Bull Bot, 2019, 54(2):265-276. |
| 杨德卫, 王莫, 韩利波, 唐定中, 李生平 . 水稻稻瘟病抗性基因的克隆、育种利用及稻瘟菌无毒基因研究进展. 植物学报, 2019,54(2):265-276. | |
| [46] | He F, Zhang H, Liu JL, Wang ZL, Wang GL . Recent advances in understanding the innate immune mechanisms and developing new disease resistance breeding strategies against the rice blast fungus Magnaporthe oryzae in rice. Hereditas(Beijing), 2014,36(8):756-765. |
| 何峰, 张浩, 刘金灵, 王志龙, 王国梁 . 水稻抗稻瘟病天然免疫机制及抗病育种新策略. 遗传, 2014,36(8):756-765. | |
| [47] | Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y . RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. Plant J, 2009,60(2):218-226. |
| [1] | Zhenwu Zheng, Hongyuan Zhao, Xiaoya Liang, Yijun Wang, Chihang Wang, Gaoyan Gong, Jinyan Huang, Guiquan Zhang, Shaokui Wang, Zupei Liu. qGL3.4 controls kernel size and plant architecture in rice [J]. Hereditas(Beijing), 2023, 45(9): 835-844. |
| [2] | Qianwen Lv, Yongfang Yang. The biological functions of peptide signaling in plant and the advances on its utilization for crop improvement [J]. Hereditas(Beijing), 2023, 45(9): 813-828. |
| [3] | Yifan Yu, Zhen OuYang, Juan Guo, Yujun Zhao, Luqi Huang. Progress on regulatory elements of plant plastid genetic engineering [J]. Hereditas(Beijing), 2023, 45(6): 501-513. |
| [4] | Fei Gao, Yu Wang, Jiaxiang Du, Xuguang Du, Jianguo Zhao, Dengke Pan, Sen Wu, Yaofeng Zhao. Advances and applications of genetically modified pig models in biomedical and agricultural field [J]. Hereditas(Beijing), 2023, 45(1): 6-28. |
| [5] | Mingliang Jiang, Hong Lang, Xiaonan Li, Ye Zu, Jing Zhao, Shenling Peng, Zhen Liu, Zongxiang Zhan, Zhongyun Piao. Progress on plant orphan genes [J]. Hereditas(Beijing), 2022, 44(8): 682-694. |
| [6] | Sanzeng Zhao, Danyu Kong, Peiyong Xin, Jinfang Chu, Yinglang Wan, Hong-Qing Ling, Yi Liu. AtCPS V326M significantly affect the biosynthesis of gibberellins [J]. Hereditas(Beijing), 2022, 44(3): 245-252. |
| [7] | Liping Tan, Rufei Gao, Xin Yin, Xuemei Chen, Fangfang Li, Liu Yuan, Junlin He. The expression of lincRNA AC027700.1 in mouse decidualization [J]. Hereditas(Beijing), 2022, 44(2): 168-177. |
| [8] | Qianbin Zhu, Zhicheng Gan, Xiaocui Li, Yingjie Zhang, Heming Zhao, Xianzhong Huang. Genome-wide identification, phylogenetic and expression of MAPKKK gene family in Arabidopsis pumila [J]. Hereditas(Beijing), 2022, 44(11): 1044-1055. |
| [9] | Yongguang Li, Yuhuan Jin, Li Guo, Hao Ai, Ruining Li, Xianzhong Huang. Genome-wide identification and expression analysis of the PEBP genes in Arabidopsis pumila [J]. Hereditas(Beijing), 2022, 44(1): 80-91. |
| [10] | Min Cao, Tongda Xu. The molecular mechanism of apical hook development in dicot plant [J]. Hereditas(Beijing), 2021, 43(8): 723-736. |
| [11] | Tianyi Wang, Yingxiang Wang, Chenjiang You. Structural and functional characteristics of plant PHD domain-containing proteins [J]. Hereditas(Beijing), 2021, 43(4): 323-339. |
| [12] | Yuanyuan Hao, Xiangqian Zhao, Fudeng Huang, Chunshou Li. The role of PPR proteins in posttranscriptional regulation of organelle components in plants [J]. Hereditas(Beijing), 2021, 43(11): 1050-1065. |
| [13] | Liping Zou, Cheng Pan, Mengxin Wang, Lin Cui, Baoyu Han. Progress on the mechanism of hormones regulating plant flower formation [J]. Hereditas(Beijing), 2020, 42(8): 739-751. |
| [14] | Xia Li, Wan Shi, Lizhao Geng, Jianping Xu. Genome editing in plants directed by CRISPR/Cas ribonucleoprotein complexes [J]. Hereditas(Beijing), 2020, 42(6): 556-564. |
| [15] | Zhichao Mei, Zhujun Wei, Jiahui Yu, Fengdan Ji, Linan Xie. Multi-omics association analysis revealed the role and mechanism of epialleles in environmental adaptive evolution of Arabidopsis thaliana [J]. Hereditas(Beijing), 2020, 42(3): 321-331. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||