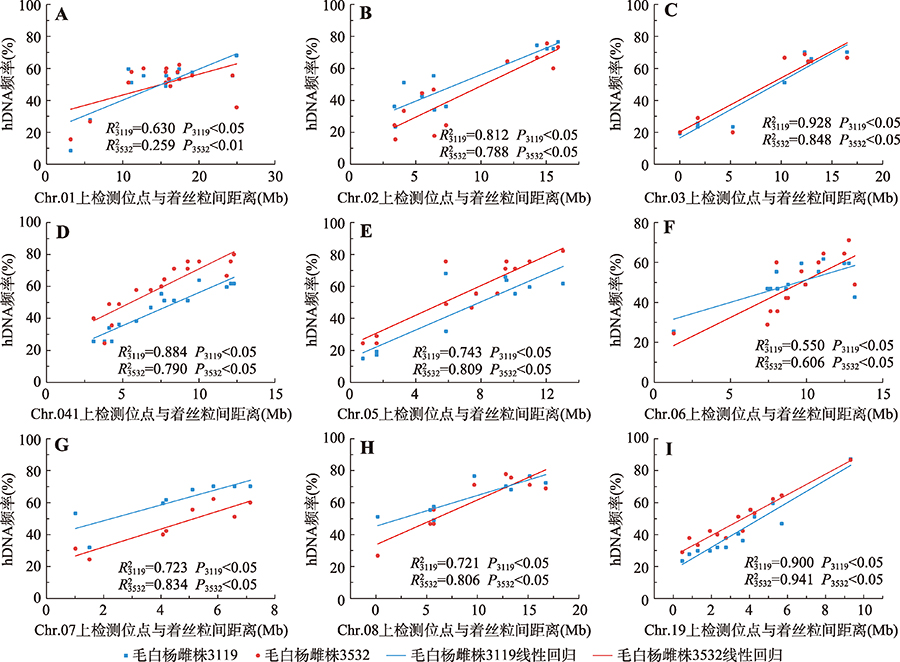
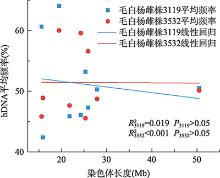
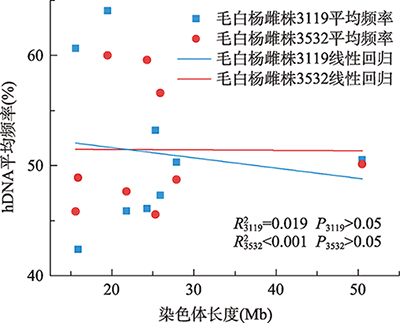
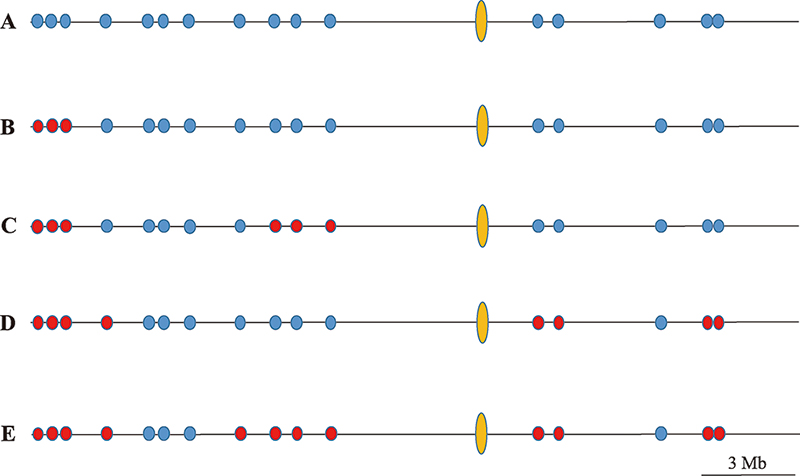
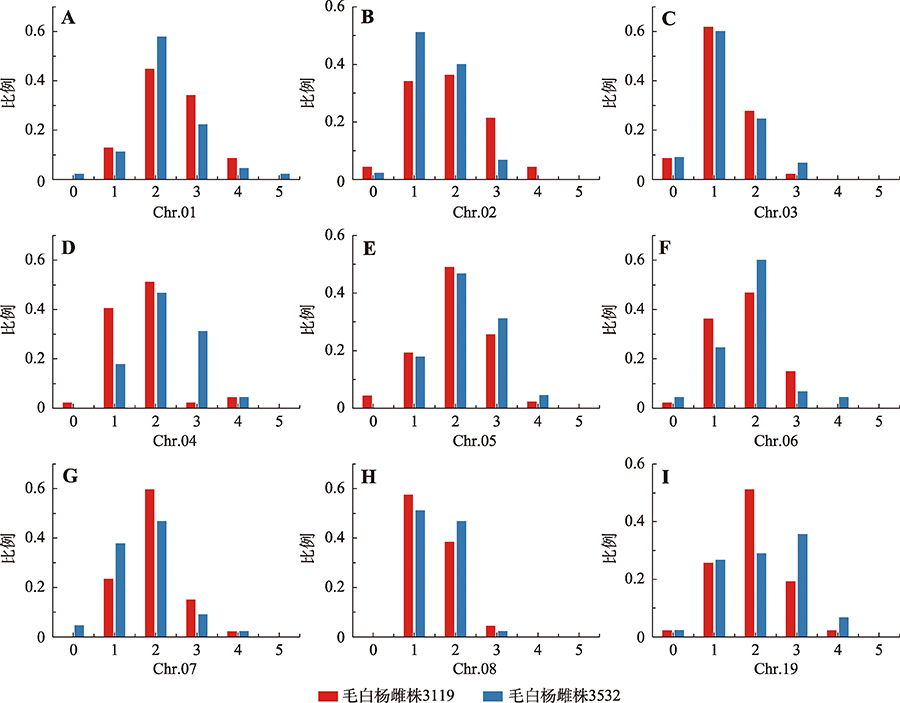
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (2): 182-193.doi: 10.16288/j.yczz.20-205
• Research article • Previous Articles Next Articles
Variation of homologous recombination in Populus tomentosa with different genotypes
Xining Geng1,2,3,4( ), Te Lu1,2,3, Kang Du1,2,3, Jun Yang1,2,3, Xiangyang Kang1,2,3(
), Te Lu1,2,3, Kang Du1,2,3, Jun Yang1,2,3, Xiangyang Kang1,2,3( )
)
- 1 Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, Beijing Forestry University, Beijing 100083, China
2 National Engineering Laboratory for Tree Breeding, Beijing Forestry University, Beijing 100083, China
3 Beijing Laboratory of Urban and Rural Ecological Environment, Beijing Forestry University, Beijing 100083, China
4 Henan Province Key Laboratory of Germplasm Innovation and Utilization of Eco-economic Woody Plant, Pingdingshan University, Pingdingshan 467000, China
-
Received:2020-07-06Online:2021-02-16Published:2021-01-22 -
Supported by:the National Natural Science Foundation of China(31470667)
Cite this article
Xining Geng, Te Lu, Kang Du, Jun Yang, Xiangyang Kang.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
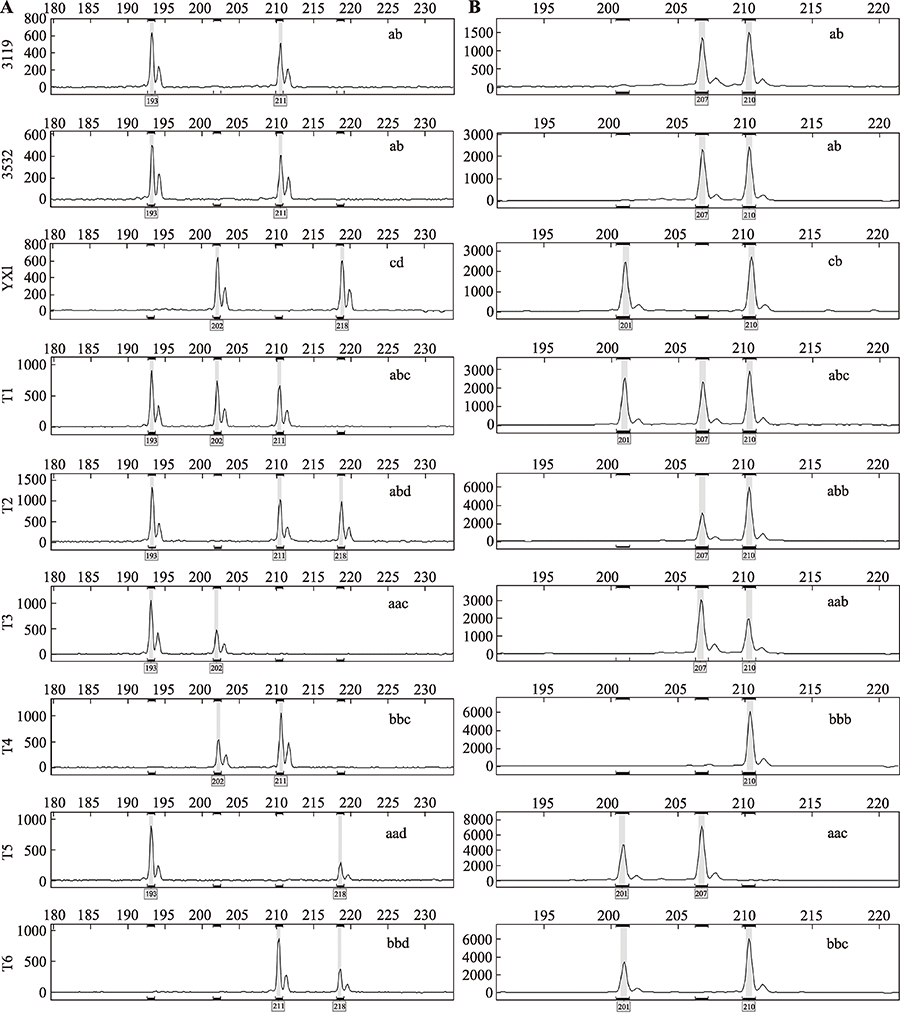
Supplemental Table 1
The information of 110 pairs of SSR primers applied for detection of hDNA"
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
|---|---|---|---|---|---|---|---|---|---|
| LG_Ⅰ-18 | 1 | 0.047 | atta | F: CACGCTAAGATTGTCTCACA | 53.35 | 45.00 | 320/332 | 320/332 | 320/328 |
| R: TCACTGCATCAAGAAGATTG | 51.30 | 40.00 | |||||||
| LG_Ⅰ-22 | 1 | 0.059 | agcag | F: AGATAAGCATGGCTTGTTGT | 51.30 | 40.00 | 309/314 | 309/314 | 309 |
| R: CGTTACCATTCATTCCAAAT | 49.25 | 35.00 | |||||||
| GCPM_2935-2 | 1 | 0.275 | ttc | F: TCAGCTTCAGCTTCTTCTTC | 53.35 | 45.00 | 192/204 | 192/204 | 198 |
| R: AATTTGAAAAGGAAAAACCC | 47.20 | 30.00 | |||||||
| LG_Ⅰ-1223 | 1 | 1.241 | gca | F: ATATCACTCGCCAAGTCAAC | 53.35 | 45.00 | 190/193 | 190/193 | 196/199 |
| R: ATCACGCACAAGTTAGGTTT | 51.30 | 40.00 | |||||||
| LG_Ⅰ-1114 | 1 | 1.424 | ta | F: AAGTGGATCAGGTGATGAAG | 53.35 | 45.00 | 156/166 | 156/166 | 165 |
| R: CAATGTCTTTCGGTTCTTTC | 51.30 | 40.00 | |||||||
| LG_Ⅰ-1065 | 1 | 1.753 | agc | F: AATGGAGGTCCCTAGCTTAC | 55.40 | 50.00 | 383/386 | 383/386 | 386 |
| R: TGATATTGCTGGGCTATCTT | 51.30 | 40.00 | |||||||
| LG_Ⅰ-1027 | 1 | 1.834 | ta | F: TGAGCTTAGGAACGAAGAAG | 53.35 | 45.00 | 188/198 | 188/198 | 184/186 |
| R: CATCAATATGACTCCTCGGT | 53.35 | 45.00 | |||||||
| LG_Ⅰ-1125 | 1 | 1.835 | tca | F: GATGCTTACCCTTTGCTCTA | 53.35 | 45.00 | 400/409 | 400/409 | 400 |
| R: CATAATGGGCAAAGAATACC | 51.30 | 40.00 | |||||||
| GCPM_3979-1 | 1 | 4.749 | ga | F: TATCAATTTCCGTGGATTTC | 49.25 | 35.00 | 192/196 | 192/196 | 193 |
| R: GAAACTGAGAGGACTTGCTG | 55.40 | 50.00 | |||||||
| GCPM_124 | 1 | 6.328 | cac | F: TTTGAGCACTTCAACTACCA | 51.30 | 40.00 | 207/210 | 207/210 | 201/210 |
| R: TGTCTTCCCTTAGTCACCAC | 55.40 | 50.00 | |||||||
| GCPM_1960-1 | 1 | 11.783 | gaaa | F: TATCGAATCACAATGGCATA | 49.25 | 35.00 | 220/224 | 220/224 | 220 |
| R: TGCTCAGGTCATCTTTTCTT | 51.30 | 40.00 | |||||||
| GCPM_3408 | 1 | 20.668 | gt | F: ATGGAAGTTTGAGTCCACTG | 53.35 | 45.00 | 202/212 | 202/212 | 200/202 |
| R: CATGCATTACTTCAACCCTT | 51.30 | 40.00 | |||||||
| GCPM_2453-1 | 1 | 28.263 | ggt | F: ACACCAAGAGCTGTAGCATT | 53.35 | 45.00 | 216/222 | 216/222 | 216/219 |
| R: ACAACATGGCCTAACTCATC | 53.35 | 45.00 | |||||||
| GCPM_1719 | 1 | 36.636 | aaag | F: AAGTGCTCATAACATCACCC | 53.35 | 45.00 | 216/228 | 216/228 | 227/234 |
| R: CTTTCCTCATTCCTGTTCTG | 53.35 | 45.00 | |||||||
| GCPM_1263-1 | 1 | 41.934 | ct | F: TGCATTAAGACATCACTTGC | 51.30 | 40.00 | 235/243 | 235/243 | 230/232 |
| R: TTCGCTTCTGTATTTCCTGT | 51.30 | 40.00 | |||||||
| PTSSR1686 | 1 | 42.469 | ttc | F: TCAGGATCGGAGTCGGAGTT | 57.45 | 55.00 | 165/168 | 165/168 | 165 |
| R: TGAGCAGAAACGACACCACT | 55.40 | 50.00 | |||||||
| GCPM_734 | 2 | 1.595 | cttt | F: GGCAATTTAGGTACAACAGC | 53.35 | 45.00 | 177/181 | 177/181 | 181 |
| R: ACAAGCGAATGCTAATTGAT | 49.25 | 35.00 | |||||||
| GCPM_2085-1 | 2 | 1.957 | ctt | F: GCAGGAACTAACAAGGTGAG | 55.40 | 50.00 | 154/157 | 154/157 | 154 |
| R: TCGGTAGCAATTGTCTGAAT | 51.30 | 40.00 | |||||||
| GCPM_3588-1 | 2 | 2.443 | aaca | F: TCGACCAACTATGTTTGACA | 51.30 | 40.00 | 184/184 | 184/184 | 191/195 |
| R: AGTTGTAACTTTGGCCTGAA | 51.30 | 40.00 |
Supplemental Table 1(Continued)
The information of 110 pairs of SSR primers applied for detection of hDNA"
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
|---|---|---|---|---|---|---|---|---|---|
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
| GCPM_1857-1 | 2 | 3.205 | tat | F: TGCCAGGGTAAGAAATAAAA | 49.25 | 35.00 | 196/212 | 196/212 | 196/200 |
| R: GAACTGAACTGATCTTGTTGC | 53.66 | 42.86 | |||||||
| ORPM_222 | 2 | 5.481 | ag | F: TGCGAACATTTTTCTTGTGG | 51.30 | 40.00 | 201/205 | 201/205 | 205 |
| R: CGCAATAGAGCCTTTGGATG | 55.40 | 50.00 | |||||||
| GCPM_2637 | 2 | 11.126 | tta | F: GACACCGTTTCTTTTCTGAG | 53.35 | 45.00 | 213/216 | 213/216 | 217 |
| R: ACCAGATCTTCATCTTCCAA | 51.30 | 40.00 | |||||||
| PMGC_2709 | 2 | 12.013 | ga | F: ATTGTAATTATTGAACACATGCC | 50.64 | 30.43 | 234/236 | 234/236 | 200/214 |
| R: GTGCAGTTCAGAGTATTGTTG | 53.66 | 42.86 | |||||||
| ORPM_161 | 2 | 13.406 | ta | F: CGGGCTAGCTCAATTAGGAG | 57.45 | 55.00 | 205/219 | 205/219 | 205/207 |
| R: ATCACCGCGAGATTCTTGTC | 55.40 | 50.00 | |||||||
| ORPM_459 | 2 | 14.121 | att | F: TTGTGGCTGTGACCAGAAAG | 55.40 | 50.00 | 227/230 | 227/230 | 233/246 |
| R: CGAGCCTCAGAAAATTGCTT | 53.35 | 45.00 | |||||||
| GCPM_2768 | 2 | 20.942 | ga | F: TTATTTGGATCCTGAAATGG | 49.25 | 35.00 | 196/206 | 186/206 | 168/200 |
| R: GATGGTTCGGTATGTGAGTT | 53.35 | 45.00 | |||||||
| PTSSR2445 | 2 | 23.932 | ag | F: CTTTGATCCCACCAACCCCA | 57.45 | 55.00 | 230/244 | 230/244 | 240/242 |
| R: TAGGCAGCTCTTTGACTGCA | 55.40 | 50.00 | |||||||
| PTSSR774 | 2 | 24.833 | tga | F: TAAGGGCAAACATGGGAGGG | 57.45 | 55.00 | 239/242 | 239/242 | 236/239 |
| R: TAGGCAGAAGTAGCAGCAGC | 57.45 | 55.00 | |||||||
| GCPM_2840-1 | 3 | 2.708 | ta | F: GAATCCAAAACAAAACATCC | 49.25 | 35.00 | 227/231 | 227/231 | 225/231 |
| R: AGGCATCAAAACTAATTGGA | 49.25 | 35.00 | |||||||
| GCPM_134-1 | 3 | 2.708 | ta | F: AGGCATCAAAACTAATTGGA | 49.25 | 35.00 | 227/230 | 227/230 | 225/231 |
| R: GAATCCAAAACAAAACATCC | 49.25 | 35.00 | |||||||
| GCPM_2628-1 | 3 | 4.469 | agc | F: TAGGCCATGTTTTTGCTTAT | 49.25 | 35.00 | 211/214 | 211/214 | 211 |
| R: GTGCCTCATTTATTGGTAGG | 53.35 | 45.00 | |||||||
| ORPM_230 | 3 | 9.739 | ga | F: CTTCCCACCCCCAACATAAC | 57.45 | 55.00 | 213/219 | 213/219 | 211/215 |
| R: TGTCATTCCACAGAGAGTCTGG | 57.67 | 50.00 | |||||||
| GCPM_1184-1 | 3 | 14.831 | ct | F: TCTTGGCGAGAGAAGTAGAG | 55.40 | 50.00 | 156/158 | 156/158 | 154/164 |
| R: GGATTTGGTGAAAATTGAAG | 49.25 | 35.00 | |||||||
| GCPM_768-1 | 3 | 16.826 | cac | F: ATTCGCTTCTTCTTCCTCTT | 51.30 | 40.00 | 208/211 | 208/211 | 211 |
| R: TATGAATTCCTCGTTGAACC | 51.30 | 40.00 | |||||||
| GCPM_2434-2 | 3 | 17.172 | tct | F: AGAGAGAGAGGTATGAGGGC | 57.45 | 55.00 | 221/224 | 221/224 | 221 |
| R: GTTCGGTAAAGGTGATGGA | 53.01 | 47.37 | |||||||
| GCPM_1468-2 | 3 | 17.480 | gac | F: CAAGTCATGGCTTCTCAAAC | 53.35 | 45.00 | 192/195 | 192/195 | 195/198 |
| R: CTATTGATCCTTGAAGACGC | 53.35 | 45.00 | |||||||
| GCPM_3465 | 3 | 20.996 | gt | F: TCAATGATTGGTCTTGTTGA | 49.25 | 35.00 | 128/134 | 128/134 | 134/146 |
| R: CAGAATTCAGAATAGAACCCA | 51.71 | 38.10 | |||||||
| GCPM_2625-1 | 4 | 0.188 | gt | F: GATTTCTATTGTGGCAAAGG | 51.30 | 40.00 | 187/193 | 187/193 | 187 |
| R: TGTATTCCTCCACTCCACTC | 55.40 | 50.00 |
Supplemental Table 1(Continued)
The information of 110 pairs of SSR primers applied for detection of hDNA"
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
|---|---|---|---|---|---|---|---|---|---|
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
| GCPM_1116-1 | 4 | 0.401 | tca | F: GGGGCTAATATCAGTTTCCT | 53.35 | 45.00 | 195/201 | 195/201 | 195 |
| R: TGAGAAAACCCTGGAAAATA | 49.25 | 35.00 | |||||||
| ORPM_394 | 4 | 0.667 | tgc | F: AAAAAGCCCCACAATTATTCA | 49.76 | 33.33 | 222/225 | 222/225 | 222/231 |
| R: GCAAGTTGCAATTGATGTCC | 53.35 | 45.00 | |||||||
| GCPM_3437 | 4 | 2.481 | ag | F: AAGATCTGGTGTTGTTTTGG | 51.30 | 40.00 | 234/236 | 234/236 | 236/242 |
| R: TTCTTAACAGAAGCCAGGAG | 53.35 | 45.00 | |||||||
| GCPM_3971-1 | 4 | 4.122 | taaa | F: CAGCCAGCCTCTTTAAGATA | 53.35 | 45.00 | 193/211 | 193/211 | 202/220 |
| R: ACAGCAATTCTGTACCTCGT | 53.35 | 45.00 | |||||||
| GCPM_1297-1 | 4 | 4.758 | tct | F: GGGATGAATCAGGAGATGTA | 53.35 | 45.00 | 135/141 | 135/141 | 140 |
| R: GAAGAAACCTGTGGGTGATA | 53.35 | 45.00 | |||||||
| ORPM_421 | 4 | 5.650 | ta | F: AAATGATGTTGCGATTTCCA | 49.25 | 35.00 | 213/221 | 213/221 | 219 |
| R: TCCCATCTCAACTACTCCAACA | 55.81 | 45.45 | |||||||
| PTSSR1738 | 4 | 6.603 | ctcttc | F: CTTCCTCGTTTGGCCCCTAA | 57.45 | 55.00 | 277/295 | 277/295 | 289/301 |
| R: ACCAGCTAGTTCGGGCTTTC | 57.45 | 55.00 | |||||||
| ORPM_201 | 4 | 7.736 | tc | F: GACTCCACCCAGTTCTGCTC | 59.50 | 60.00 | 214/226 | 214/226 | 224/226 |
| R: AACTTCCCATCGAATGATCG | 53.35 | 45.00 | |||||||
| PTSSR1873 | 4 | 8.381 | GA | F: GCGTGAGATGAGGGAGAGTG | 59.50 | 60.00 | 184/198 | 184/198 | 186/202 |
| R: CGTCAGGCGACAATTCCAAC | 57.45 | 55.00 | |||||||
| PTSSR1932 | 4 | 9.402 | TAC | F: CGGCAGGTCTGATGTCTTCA | 57.45 | 55.00 | 120/123 | 120/123 | 117/123 |
| R: CCATGGAGTCTAAAGGTGGCA | 57.57 | 52.38 | |||||||
| GCPM_3819-1 | 4 | 16.315 | gaa | F: ATCATCAAGTTCACGAAAGC | 51.30 | 40.00 | 240/243 | 240/243 | 237 |
| R: ACTGACCTACCACCATTTTG | 53.35 | 45.00 | |||||||
| GCPM_295-1 | 4 | 16.784 | aag | F: TAAGAGGATTACCAGGCAGA | 53.35 | 45.00 | 136/145 | 136/145 | 142/148 |
| R: CCCACTAACAATAAGCTTGG | 53.35 | 45.00 | |||||||
| PTSSR1623 | 4 | 20.046 | tga | F: CATCTCCCACCACCACAACA | 57.45 | 55.00 | 238/248 | 238/248 | 238 |
| R: CTCTGCGTAAACAACTGCGG | 57.45 | 55.00 | |||||||
| PTSSR2267 | 4 | 21.767 | tca | F: TGGAAGGCAAATCGTCGTCA | 55.40 | 50.00 | 182/197 | 182/197 | 190 |
| R: AAGTGGAGCAGACAGACACT | 55.40 | 50.00 | |||||||
| GCPM_3234-1 | 4 | 21.772 | agc | F: TCTCAATGTCGACACCATTA | 51.30 | 40.00 | 195/198 | 195/198 | 195 |
| R: TGCAAGAACTGCTATCACTG | 53.35 | 45.00 | |||||||
| GCPM_1255 | 5 | 1.974 | ag | F: GAACCTTAAAACCAGAACCC | 53.35 | 45.00 | 191/193 | 191/193 | 189/193 |
| R: GAGCCACAGAAATACTGCTC | 55.40 | 50.00 | |||||||
| GCPM_379-1 | 5 | 4.025 | tat | F: GGCCATTCATTAGATGTAGC | 53.35 | 45.00 | 153/156 | 153/156 | 153 |
| R: AAATCAAGATCCAAAGCAAA | 47.20 | 30.00 | |||||||
| GCPM_1063 | 5 | 5.996 | ca | F: AGTTAATTGCGCATGTTCTT | 49.25 | 35.00 | 168/188 | 168/188 | 170/176 |
| R: AAACAAACTCCAGCAAACAT | 49.25 | 35.00 | |||||||
| GCPM_3536-2 | 5 | 9.139 | ttg | F: AGATTCTTTTTCGGCTTCTT | 49.25 | 35.00 | 137/146 | 137/146 | 141/144 |
| R: AGAAGATGCTGGAGTTCAGA | 53.35 | 45.00 | |||||||
| GCPM_1838 | 5 | 9.156 | ga | F: GTTCAGCGAAAGCTAAAGAG | 53.35 | 45.00 | 142/146 | 142/146 | 138/168 |
| R: CACAGAATTACAGCTGATGC | 53.35 | 45.00 |
Supplemental Table 1(Continued)
The information of 110 pairs of SSR primers applied for detection of hDNA"
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
|---|---|---|---|---|---|---|---|---|---|
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
| GCPM_2013-1 | 5 | 14.238 | At | F: TTTAGGGTCGTAGTGGAAAA | 51.30 | 40.00 | 238/240 | 238/240 | 250/254 |
| R: AAACCTGGAGGAGAATTAGAA | 51.71 | 38.10 | |||||||
| ORPM_136 | 5 | 16.608 | ct | F: TTTAAGCCTCCGAAAACCAA | 51.30 | 40.00 | 228/240 | 228/240 | 232 |
| R: CTGCAAGGCGAGGTTATTCT | 55.40 | 50.00 | |||||||
| ORPM_442 | 5 | 16.608 | ct | F: TGGTTTTCGGAGGCTTAAAA | 51.30 | 40.00 | 244/254 | 244/254 | 236 |
| R: CCTGCAGCGAGTATTAATTGG | 55.61 | 47.62 | |||||||
| GCPM_3151 | 5 | 22.434 | ag | F: ACCATCATTAACCCCACATA | 51.30 | 40.00 | 144/156 | 144/156 | 135/155 |
| R: AAAGAAACCAGACCACACAC | 53.35 | 45.00 | |||||||
| GCPM_2858-1 | 5 | 22.702 | tg | F: TGCAAGTCTTTTTAGGAACC | 51.30 | 40.00 | 254/256 | 254/256 | 254/260 |
| R: TTCAAAATGCATCAAAGTGT | 47.20 | 30.00 | |||||||
| GCPM_540-1 | 5 | 24.498 | taa | F: GATGGGGAGGTTATTTTCTT | 51.30 | 40.00 | 154/160 | 154/160 | 154 |
| R: CAATATTGAGGAAAATCAAAGG | 50.22 | 31.82 | |||||||
| GCPM_4008-1 | 5 | 24.561 | acc | F: AGAGAGAAGCTTGTGTCCAG | 55.40 | 50.00 | 161/170 | 161/170 | 170 |
| R: TGAGGAAGCAGAAGTAGAGC | 55.40 | 50.00 | |||||||
| ORPM_25 | 5 | 25.091 | ta | F: AAGAGTTGAAGGCTGGACGA | 55.40 | 50.00 | 236/240 | 236/240 | 236 |
| R: AGACATGCATGAAGCCATGA | 53.35 | 45.00 | |||||||
| GCPM_547-1 | 6 | 1.831 | at | F: CCTCTTGAAAAGAAGCAAAA | 49.25 | 35.00 | 217/231 | 217/231 | 213 |
| R: ATCAAAAATGCCGATTAAAA | 45.15 | 25.00 | |||||||
| GCPM_139 | 6 | 2.516 | gt | F: ATGACATGACATGATTGGAA | 49.25 | 35.00 | 220/228 | 220/228 | 200/226 |
| R: CTTCTGCTGGAAGAAGAAAA | 51.30 | 40.00 | |||||||
| GCPM_2126 | 6 | 3.888 | ag | F: CACGTAAACAGCTTCCAAGT | 53.35 | 45.00 | 178/194 | 178/194 | 160/176 |
| R: TAATGATTCCAGCTATGGGT | 51.30 | 40.00 | |||||||
| GCPM_1072-2 | 6 | 4.204 | ttta | F: AGGAAAACAAAGGAGAGGAG | 53.35 | 45.00 | 150/154 | 150/154 | 142/154 |
| R: ATGCTTAAAAGGGGATCTCT | 51.30 | 40.00 | |||||||
| GCPM_3539-2 | 6 | 5.331 | aat | F: TATTCGGTACAAGACTTGGG | 53.35 | 45.00 | 215/218 | 215/218 | 212/215 |
| R: TAATTGTAAAGCGGCTATCG | 51.30 | 40.00 | |||||||
| GCPM_2642-2 | 6 | 6.965 | cca | F: GCTTAGCTGGATGAGAAGAA | 53.35 | 45.00 | 239/241 | 239/241 | 239 |
| R: ATAGTTACAGGCCACCATTG | 53.35 | 45.00 | |||||||
| PTSSR580 | 6 | 13.716 | atg | F: AATGCATCCTCAGCTCCAGG | 57.45 | 55.00 | 281/284 | 281/284 | 281 |
| R: CGGCGGCTGCAAACATTAAT | 55.40 | 50.00 | |||||||
| GCPM_1017-1 | 6 | 22.438 | ataa | F: GTTTAATTCCCACGTCGTTA | 51.30 | 40.00 | 171/175 | 171/175 | 175/177 |
| R: CGAATGAAGAAAAACCATTC | 49.25 | 35.00 | |||||||
| GCPM_2705-1 | 6 | 22.633 | ac | F: TTTGCACAGGTAAGTTGATG | 51.30 | 40.00 | 146/150 | 146/150 | 148/152 |
| R: ATTGACCATAGCAGACAACC | 53.35 | 45.00 | |||||||
| GCPM_2034 | 6 | 23.087 | ag | F: ACAAACTGCTTTGTTTGGTT | 49.25 | 35.00 | 176/190 | 176/190 | 186/196 |
| R: CTCCATTCATAAAATCGAGC | 51.30 | 40.00 | |||||||
| GCPM_1632-1 | 6 | 23.663 | ct | F: TTTCTCTCTCTGAAACCCCT | 53.35 | 45.00 | 211/225 | 211/225 | 203/221 |
| R: AGCGACTCACTGAGCTTTAG | 55.40 | 50.00 | |||||||
| ORPM_365 | 6 | 23.788 | tg | F: GGGTTGGACCTGCTCAAATA | 55.40 | 50.00 | 226/228 | 226/228 | 226/238 |
| R: CAGCTGCTTTTTCATGGCTA | 53.35 | 45.00 |
Supplemental Table 1(Continued)
The information of 110 pairs of SSR primers applied for detection of hDNA"
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
|---|---|---|---|---|---|---|---|---|---|
| GCPM_2627-1 | 6 | 24.930 | cac | F: TAAGTCCCACTACACCCAAC | 55.40 | 50.00 | 234/254 | 234/254 | 240/255 |
| R: GAGTTCGAGAGAGGGAATCT | 55.40 | 50.00 | |||||||
| GCPM_2615-1 | 6 | 27.782 | cttt | F: ATGTCAACGTCACTGACAAA | 51.30 | 40.00 | 231/237 | 231/237 | 229/236 |
| R: ATTAGGCAATGCAGAACACT | 51.30 | 40.00 | |||||||
| GCPM_1054-1 | 7 | 0.863 | ct | F: AGGTCTGTGCAAGGAATAAA | 51.30 | 40.00 | 159/165 | 159/165 | 154/165 |
| R: GTCTGTAATCAAGCCAAAGC | 53.35 | 45.00 | |||||||
| GCPM_741-1 | 7 | 1.416 | at | F: CCGTTTGATTAAAAAGATGC | 49.25 | 35.00 | 212/214 | 212/214 | 214 |
| R: TTATTGAGCTGATGATCCCT | 51.30 | 40.00 | |||||||
| WPMS_17 | 7 | 6.495 | cac | F: ACATCCGCCAATGCTTCGGTGTTT | 59.57 | 50.00 | 137/146 | 137/146 | 137/140 |
| R: GTGACGGTGGTGGCGGATTTTCTT | 61.28 | 54.17 | |||||||
| GCPM_3738-1 | 7 | 9.014 | ac | F: TGAACAAGACACCAAAATGA | 49.25 | 35.00 | 133/139 | 133/139 | 133 |
| R: TCTCGACTTTACCATCTCGT | 53.35 | 45.00 | |||||||
| GCPM_2741-1 | 7 | 12.076 | at | F: CAAGCAGTATCTTCCACTGA | 53.35 | 45.00 | 142/146 | 142/148 | 134 |
| R: TAGCCAACCACTCTAAAGGA | 53.35 | 45.00 | |||||||
| GCPM_3476-1 | 7 | 12.189 | ggt | F: GGGAATGTAAGGATGTGTTG | 53.35 | 45.00 | 202/220 | 202/220 | 205/211 |
| R: AACCAGAAAACGACAGTCAC | 53.35 | 45.00 | |||||||
| GCPM_3332-1 | 7 | 13.116 | ag | F: TCCACTGCCTATGAACTTTT | 51.30 | 40.00 | 138/154 | 138/154 | 138 |
| R: CACCCAATAGCTTCCATATT | 51.30 | 40.00 | |||||||
| GCPM_3727-1 | 7 | 13.848 | ct | F: TTGGGGTTAGTGACTAGTGG | 55.40 | 50.00 | 152/158 | 152/158 | 152/154 |
| R: CAAGCTGTGTAAAGACACCA | 53.35 | 45.00 | |||||||
| LG_Ⅷ-50 | 8 | 0.255 | ga | F: AACAACAGATCGCTGAAACT | 51.30 | 40.00 | 265/281 | 265/281 | 265/271 |
| R: CCATGTACTTAGGCATGGAT | 53.35 | 45.00 | |||||||
| GCPM_2992 | 8 | 1.833 | TA | F: GATCATAACCCAGGAAACAA | 51.30 | 40.00 | 237/241 | 237/241 | 241 |
| R: CGTGAATTTTTGGGATTTTA | 47.20 | 30.00 | |||||||
| GCPM_1344-2 | 8 | 3.669 | tat | F: GAGACCTGAGAAACAGAGGA | 55.40 | 50.00 | 227/230 | 227/230 | 230 |
| R: CAAATGCAACGCAATAATAA | 47.20 | 30.00 | |||||||
| GCPM_33-1 | 8 | 4.192 | tta | F: TTTGGCCTTAACTCTCCATA | 51.30 | 40.00 | 171/174 | 171/174 | 167/179 |
| R: AGTTGCTCTCAAAGAAGGTT | 51.30 | 40.00 | |||||||
| GCPM_13-1 | 8 | 7.309 | taa | F: ATTGTTCTTGTTGAAGGACG | 51.30 | 40.00 | 207/213 | 207/213 | 204/210 |
| R: AGAGCAAACAAATTGATGGT | 49.25 | 35.00 | |||||||
| ORPM_126 | 8 | 11.293 | [ct]4cct | F: GCCGAAGTTGACGATAGCTC | 57.45 | 55.00 | 215/222 | 215/222 | 215 |
| R: GAGTTAAACCCACCCTGCAA | 55.40 | 50.00 | |||||||
| ORPM_455 | 8 | 11.293 | ga | F: GAGTTAAACCCACCCTGCAA | 55.40 | 50.00 | 215/225 | 215/225 | 215 |
| R: GCCGAAGTTGACGATAGCTC | 57.45 | 55.00 | |||||||
| GCPM_2958-1 | 8 | 11.639 | ct | F: AAAGAGGATTACTTAGGCGG | 53.35 | 45.00 | 201/215 | 201/215 | 217/219 |
| R: ACGAGAACATTCATCAATCC | 51.30 | 40.00 | |||||||
| GCPM_3474-1 | 8 | 16.814 | agc | F: GATCCGAAAACAACAACAAT | 49.25 | 35.00 | 123/135 | 123/135 | 123 |
| R: ACCCCTTTCTCTTCTCAATC | 53.35 | 45.00 |
Supplemental Table 1(Continued)
The information of 110 pairs of SSR primers applied for detection of hDNA"
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
|---|---|---|---|---|---|---|---|---|---|
| 标记名称 | 染色 体 | 位置 (Mb) | 重复 单元 | 引物序列(5?→3?) | Tm (℃) | GC含 量(%) | 3532中的 等位基因(bp) | 3119中的 等位基因(bp) | YX1中的 等位基因(bp) |
| MB50065 | 19 | 0.301 | A | F: CACCAAAATTAAAGGGCCAA | 204/218 | 204/218 | 201 | ||
| R: GCATGCTAGTGCCTTTTCCT | |||||||||
| MB41376 | 19 | 1.940 | at | F: GAGCAGGTGACTCGTGAACA | 251/255 | 251/255 | 251 | ||
| R: GGAGAGTGATAAAACCAACAATCC | |||||||||
| MB41583 | 19 | 2.580 | gcg | F: CAAGTTGGCTTGGATGGATT | 144/162 | 144/162 | 146/149 | ||
| R: AACCAGCCCAACAAATCAAG | |||||||||
| MB55349 | 19 | 3.680 | tc | F: GGACACGTGATGTCATGGAG | 264/268 | 264/268 | 258/260 | ||
| R: TCAAATGCAAAGCCCTTTTT | |||||||||
| MB54907 | 19 | 4.713 | ga | F: TAGCAAAAGCAGCAGCAGAG | 262/300 | 262/300 | 262/378 | ||
| R: CAAGTGCGTGAAAAACATGG | |||||||||
| MB54763 | 19 | 5.159 | tta | F: AGTTGAATTTGCGCTTCGAT | 128/134 | 128/134 | 128 | ||
| R: TAGGGTGTGGCCAACATACA | |||||||||
| MB84322 | 19 | 5.526 | tta | F: GCATGCTGTTTGCATCTAAAG | 234/248 | 234/248 | 234 | ||
| R: ATTAAACCTCGGGTCCTGGT | |||||||||
| MB120414 | 19 | 7.927 | tg | F: GTGGCGAAGCTTTATTCCTG | 297/311 | 297/311 | 307/311 | ||
| R: TAAAGCCCAAAGCAGTCACC | |||||||||
| LG_XIX-11 | 19 | 8.770 | atca | F: CAACATGAAATGAGCTGCTA | 398/404 | 398/404 | 404/408 | ||
| R: TCCACATGATGTCTGATTTG | |||||||||
| MB125350 | 19 | 9.649 | tc | F: AAAGAGTCAACGGGCAAAAA | 284/290 | 284/290 | 292/300 | ||
| R: AGTGGCAGCTGAATGGAAAT | |||||||||
| MB37553 | 19 | 10.266 | ta | F: TGACCAAGAGATTGCCACAG | 298/306 | 298/306 | 317/343 | ||
| R: GAAAGTTTCATGTGCCGGAT | |||||||||
| MB125779 | 19 | 11.237 | t | F: TTGAAGTATGATTAGCATCCTCG | 112/115 | 112/115 | 115/123 | ||
| R: TCCCTCTGGTTACCATTTGC | |||||||||
| MB70130 | 19 | 15.319 | a | F: TGCTCCAGAGGTTACTCCGT | 233/239 | 233/239 | 233 | ||
| R: TCCAATGATGGATCTGGTGA |
| [1] |
Cromie GA, Smith GR. Branching out: meiotic recombination and its regulation. Trends Cell Biol , 2007, 17(9): 448-455. Homologous recombination is a dynamic process by which DNA sequences and strands are exchanged. In meiosis, the reciprocal DNA recombination events called crossovers are central to the generation of genetic diversity in gametes and are required for homolog segregation in most organisms. Recent studies have shed light on how meiotic crossovers and other recombination products form, how their position and number are regulated and how the DNA molecules undergoing recombination are chosen. These studies indicate that the long-dominant, unifying model of recombination proposed by Szostak et al. applies, with modification, only to a subset of recombination events. Instead, crossover formation and its control involve multiple pathways, with considerable variation among model organisms. These observations force us to ‘branch out’ in our thinking about meiotic recombination.
doi: 10.1016/j.tcb.2007.07.007 |
| [2] | Melamed-Bessudo C, Shilo S, Levy AA. Meiotic recombination and genome evolution in plants. Curr Opin Plant Biol , 2016, 30: 82-87. |
| [3] |
Mancera E, Bourgon R, Huber W, Steinmetz LM. Genome-wide survey of post-meiotic segregation during yeast recombination. Genome Biol , 2011, 12(4): R36.
doi: 10.1186/gb-2011-12-4-r36 pmid: 21481229 |
| [4] |
Martini E, Borde V, Legendre M, Audic S, Regnault B, Soubigou G, Dujon B, Llorente B. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet , 2011, 7(9): e1002305.
doi: 10.1371/journal.pgen.1002305 pmid: 21980306 |
| [5] |
Preuss D, Rhee SY, Davis RW. Tetrad analysis possible in Arabidopsis with mutation of the quartet ( qrt ) genes . Science , 1994, 264(5164): 1458-1460.
pmid: 8197459 |
| [6] | Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, Copenhaver GP. Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis . Proc Natl Acad Sci USA , 2007, 104(10): 3913-3918. |
| [7] | Sun YJ, Ambrose JH, Haughey BS, Webster TD, Pierrie SN, Muñoz DF, Wellman EC, Cherian S, Lewis SM, Berchowitz LE, Copenhaver GP. Deep genome-wide measurement of meiotic gene conversion using tetrad analysis in Arabidopsis thaliana . PLoS Genet , 2012, 8(10): e1002968. |
| [8] |
Berchowitz LE, Copenhaver GP. Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets . Nat Protoc , 2008, 3(1): 41-50.
doi: 10.1038/nprot.2007.491 pmid: 18193020 |
| [9] |
Yelina NE, Ziolkowski PA, Miller N, Zhao XH, Kelly KA, Muñoz DF, Mann DJ, Copenhaver GP, Henderson IR. High-throughput analysis of meiotic crossover frequency and interference via flow cytometry of fluorescent pollen in Arabidopsis thaliana . Nat Protoc , 2013, 8(11): 2119-2134.
doi: 10.1038/nprot.2013.131 pmid: 24113785 |
| [10] |
Li X, Li L, Yan JB. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat Commun , 2015, 6: 6648.
pmid: 25800954 |
| [11] |
Dreissig S, Fuchs J, Himmelbach A, Mascher M, Houben A. Sequencing of single pollen nuclei reveals meiotic recombination events at megabase resolution and circumvents segregation distortion caused by postmeiotic processes . Front Plant Sci , 2017, 8: 1620.
doi: 10.3389/fpls.2017.01620 pmid: 29018459 |
| [12] | Dong CB, Mao JF, Suo YJ, Shi L, Wang J, Zhang PD, Kang XY. A strategy for characterization of persistent heteroduplex DNA in higher plants. Plant J , 2014, 80(2): 282-291. |
| [13] | Bauer E, Falque M, Walter H, Bauland C, Camisan C, Campo L, Meyer N, Ranc N, Rincent R, Schipprack W, Altmann T, Flament P, Melchinger AE, Menz M, Moreno-González J, Ouzunova M, Revilla P, Charcosset A, Martin OC, Schön CC. Intraspecific variation of recombination rate in maize. Genome Biol , 2013, 14(9): R103. |
| [14] |
Yin TM, Difazio SP, Gunter LE, Zhang XY, Sewell MM, Woolbright SA, Allan GJ, Kelleher CT, Douglas CJ, Wang MX, Tuskan GA. Genome structure and emerging evidence of an incipient sex chromosome in Populus . Genome Res , 2008, 18(3): 422-430.
doi: 10.1101/gr.7076308 pmid: 18256239 |
| [15] | Kersten B, Pakull B, Fladung M. Genomics of sex determination in dioecious trees and woody plants. Trees , 2017, 31(4): 1113-1125. |
| [16] | Kang N, Bai FY, Zhang PD, Kang XY. Inducing chromosome doubling of embryo sac in Populus tomentosa with high temperature exposure for hybrid triploids . J Beijing Fore Univ , 2015, 37(2): 79-86. |
| 康宁, 白凤莹, 张平冬, 康向阳. 高温诱导胚囊染色体加倍获得毛白杨杂种三倍体. 北京林业大学学报, 2015, 37(2): 79-86. | |
| [17] | Li YH. Chromosome doubling of female gametes in white poplars[Dissertation]. Beijing Forstry University, 2007. |
| 李艳华. 白杨雌配子染色体加倍技术研究[学位论文]. 北京林业大学, 2007. | |
| [18] | Wang J. Techniques of polyploid induction in Populus spp. (Section Tacamahaca )[Dissertation]. Beijing Forstry University, 2009. |
| 王君. 青杨派树种多倍体诱导技术研究[学位论文]. 北京林业大学, 2009. | |
| [19] |
Schuelke M. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol , 2000, 18(2): 233-234.
doi: 10.1038/72708 pmid: 10657137 |
| [20] | Hulce D, Li X, Snyder-Leiby T, Johathan LC. GeneMarker® Genotyping Software: Tools to increase the statistical power of dna fragment analysis. J Biomol Tech , 2011,22(Suppl.): S35-S36. |
| [21] |
Yin TM, Zhang XY, Gunter LE, Li SX, Wullschleger SD, Huang MR, Tuskan GA. Microsatellite primer resource for Populus developed from the mapped sequence scaffolds of the Nisqually-1 genome . New Phytol , 2009, 181(2): 498-503.
doi: 10.1111/j.1469-8137.2008.02663.x pmid: 19121044 |
| [22] |
Vining KJ, Pomraning KR, Wilhelm LJ, Priest HD, Pellegrini M, Mockler TC, Freitag M, Strauss SH. Dynamic DNA cytosine methylation in the Populus trichocarpa genome: tissue-level variation and relationship to gene expression . BMC Genomics , 2012, 13: 27.
doi: 10.1186/1471-2164-13-27 pmid: 22251412 |
| [23] |
Xin HY, Zhang T, Han YH, Wu YF, Shi JS, Xi ML, Jiang JM. Chromosome painting and comparative physical mapping of the sex chromosomes in Populus tomentosa and Populus deltoides . Chromosoma , 2018, 127(3): 313-321.
doi: 10.1007/s00412-018-0664-y pmid: 29520650 |
| [24] | Si WN, Yuan Y, Huang J, Zhang XH, Zhang YC, Zhang YD, Tian DC, Wang CL, Yang YH, Yang SH. Widely distributed hot and cold spots in meiotic recombination as shown by the sequencing of rice F 2 plants . New Phytol , 2015, 206(4): 1491-1502. |
| [25] |
Wijnker E, Velikkakam James G, Ding J, Becker F, Klasen JR, Rawat V, Rowan BA, de Jong DF, de Snoo CB, Zapata L, Huettel B, de Jong H, Ossowski S, Weigel D, Koornneef M, Keurentjes JJ, Schneeberger K. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana . eLife , 2013, 2: e01426.
doi: 10.7554/eLife.01426 pmid: 24347547 |
| [26] |
Henderson IR. Control of meiotic recombination frequency in plant genomes. Curr Opin Plant Biol , 2012, 15(5): 556-561.
pmid: 23017241 |
| [27] |
Yelina N, Diaz P, Lambing C, Henderson IR. Epigenetic control of meiotic recombination in plants. Sci China Life Sci , 2015, 58(3): 223-231.
doi: 10.1007/s11427-015-4811-x pmid: 25651968 |
| [28] |
Kleckner N, Storlazzi A, Zickler D. Coordinate variation in meiotic pachytene SC length and total crossover/ chiasma frequency under conditions of constant DNA length. Trends Genet , 2003, 19(11): 623-628.
doi: 10.1016/j.tig.2003.09.004 pmid: 14585614 |
| [29] |
Ruiz-Herrera A, Vozdova M, Fernández J, Sebestova H, Capilla L, Frohlich J, Vara C, Hernández-Marsal A, Sipek J, Robinson TJ, Rubes J. Recombination correlates with synaptonemal complex length and chromatin loop size in bovids-insights into mammalian meiotic chromosomal organization. Chromosoma , 2017, 126(5): 615-631.
doi: 10.1007/s00412-016-0624-3 pmid: 28101670 |
| [30] | Quevedo C, Del Cerro AL, Santos JL, Jones GH. Correlated variation of chiasma frequency and synaptonemal complex length in Locusta migratoria . Heredity , 1997, 78(5): 515-519. |
| [31] | Lynn A, Koehler KE, Judis L, Chan ER, Cherry JP, Schwartz S, Seftel A, Hunt PA, Hassold TJ. Covariation of synaptonemal complex length and mammalian meiotic exchange rates. Science , 2002, 296(5576): 2222-2225. |
| [32] |
Anderson LK, Stack SM, Fox MH, Zhang CS. The relationship between genome size and synaptonemal complex length in higher plants. Exp Cell Res , 1985, 156(2): 367-378.
doi: 10.1016/0014-4827(85)90544-0 pmid: 3967684 |
| [33] |
Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes . PLoS Genet , 2011, 7(11): e1002354.
doi: 10.1371/journal.pgen.1002354 pmid: 22072983 |
| [34] | Basheva EA, Bidau CJ, Borodin PM. General pattern of meiotic recombination in male dogs estimated by MLH1 and RAD51 immunolocalization. Chromosome Res , 2008, 16(5): 709-719. |
| [35] | Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non- crossovers in yeast. Nature , 2008, 454(7203): 479-485. |
| [36] |
Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science , 2003, 301(5634): 785-789.
pmid: 12907787 |
| [37] |
Jones GH. The control of chiasma distribution. Symp Soc Exp Biol , 1984, 38: 293-320.
pmid: 6545727 |
| [38] |
Ritz KR, Noor MAF, Singh ND. Variation in recombination rate: adaptive or not? Trends Genet , 2017, 33(5): 364-374.
doi: 10.1016/j.tig.2017.03.003 pmid: 28359582 |
| [39] |
Drouaud J, Mercier R, Chelysheva L, Bérard A, Falque M, Martin O, Zanni V, Brunel D, Mézard C. Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4 . PLoS Genet , 2007, 3(6): e106.
doi: 10.1371/journal.pgen.0030106 pmid: 17604455 |
| [40] |
Luo C, Li X, Zhang QH, Yan JB. Single gametophyte sequencing reveals that crossover events differ between sexes in maize. Nat Commun , 2019, 10(1): 785.
pmid: 30770831 |
| [41] | Dong CB. Analysis of homologous recombination characteristics and 2n gamete heterozygosity in Populus [Dissertation]. Beijing Forestry University, 2015. |
| 董春波. 杨树同源重组特点及2n配子杂合性分析[学位论文]. 北京林业大学, 2015. | |
| [42] | Yang SH, Yuan Y, Wang L, Li J, Wang W, Liu HX, Chen JQ, Hurst LD, Tian DC. Great majority of recombination events in Arabidopsis are gene conversion events . Proc Natl Acad Sci USA , 2012, 109(51): 20992-20997. |
| [43] | Hadad RG, Pfeiffer TW, Poneleit CG. Repeatability and heritability of divergent recombination frequencies in the Iowa Stiff Stalk Synthetic ( Zea mays L. ). Theor Appl Genet , 1996, 93( 5-6): 990-996. |
| [1] | Yuxuan Guo, Shunping Yan, Yingxiang Wang. Recent advances in functional conservation and divergence of recombinase RAD51 and DMC1 [J]. Hereditas(Beijing), 2022, 44(5): 398-413. |
| [2] |
Yanyan Cao, Miaomiao Cheng, Fang Song, Yujin Qu, Jinli Bai, Hong Wang.
|
| [3] | Fan Li, Rongpei Yu, Dan Sun, Jihua Wang, Shenchong Li, Jiwei Ruan, Qinli Shan, Pingli Lu, Guoxian Wang. Molecular mechanisms of meiotic recombination suppression in plants [J]. Hereditas(Beijing), 2019, 41(1): 52-65. |
| [4] | Quan Rong, Li Guoling, Mo Jianxin, Zhong Cuili, Li Zicong, Gu Ting, Zheng Enqin, Liu Dewu, Cai Gengyuan, Wu Zhenfang, Zhang Xianwei. Effects of RNA interference on the porcine NHEJ pathway repair factors on HR efficiency [J]. Hereditas(Beijing), 2018, 40(9): 749-757. |
| [5] | Caijiao Liang, Fanmei Meng, Yuncan Ai. CRISPR/Cas systems in genome engineering of bacteriophages [J]. Hereditas(Beijing), 2018, 40(5): 378-389. |
| [6] | Chunxia Liu, Lizhao Geng, Jianping Xu. Detection methods of genome editing in plants [J]. Hereditas(Beijing), 2018, 40(12): 1075-1091. |
| [7] |
Min Huang,Yeran Yang,Xiaoyan Sun,Ting Zhang,Caixia Guo.
RAD51 regulates REV1 recruitment to DNA double-strand break sites [J]. Hereditas(Beijing), 2018, 40(11): 1007-1014. |
| [8] | Yan He,Mengnv Xie,Li Yu,Zhen Ren,Fang Zhu,Chun Fu. The roles of Fanconi anemia genes in the regulation of follicle development [J]. Hereditas(Beijing), 2017, 39(6): 469-481. |
| [9] | Xiaoling Zhang,Yun Long,Fei Ge,Zhongrong Guan,Xiaoxiang Zhang,Yanli Wang,Yaou Shen,Guangtang Pan. A genetic study of the regeneration capacity of embryonic callus from the maize immature embryo culture [J]. Hereditas(Beijing), 2017, 39(2): 143-155. |
| [10] | Guoling Li,Cuili Zhong,Jianxin Mo,Rong Quan,Zhenfang Wu,Zicong Li,Huaqiang Yang,Xianwei Zhang. Advances in site-specific integration of transgene in animal genome [J]. Hereditas(Beijing), 2017, 39(2): 98-109. |
| [11] | Wenchao Zou, Linlin Shen, Jianguo Shen, Wei Cai, Jiasui Zhan, Fangluan Gao. A multigene phylogentic analysis of Potato virus Y and its application in strain identification [J]. Hereditas(Beijing), 2017, 39(10): 918-929. |
| [12] | Wei Wang, Yushuang Wang, Lanlan Huang, Zijian Jian, Xinhua Wang, Shouren Liu, Wenhui Pi. Increasing the efficiency of homologous recombination vector-mediated end joining repair by inhibition of Lig4 gene using siRNA in sheep embryo fibroblasts [J]. Hereditas(Beijing), 2016, 38(9): 831-839. |
| [13] | Xianwei Yang, Ruifu Yang, Yujun Cui. Homologous recombination among bacterial genomes: the measurement and identification [J]. HEREDITAS(Beijing), 2016, 38(2): 137-143. |
| [14] | Xuli Qian, Xin Cao. Phenotype predictions of the pathogenic nonsynonymous single nucleotide polymorphisms in deafness-causing gene COCH [J]. HEREDITAS(Beijing), 2015, 37(7): 664-672. |
| [15] | Xiaoli Wang,Chuang Jiang,Jianhua Liu,Xipeng Liu. An efficient genetic knockout system based on linear DNA fragment homologous recombination for halophilic archaea [J]. HEREDITAS(Beijing), 2015, 37(4): 388-395. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||