Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (3): 226-239.doi: 10.16288/j.yczz.20-268
Previous Articles Next Articles
Progress on animal domestication under population genetics
Zilong Wen1( ), Yiqiang Zhao1(
), Yiqiang Zhao1( )
)
- 1 College of Biological sciences, China Agricultural University, Beijing 100193, China
-
Received:2020-08-26Online:2021-03-16Published:2021-02-01 -
Supported by:National Special Foundation for Transgenic Species of China(2018ZX08007001)
Cite this article
Zilong Wen, Yiqiang Zhao. Progress on animal domestication under population genetics[J]. Hereditas(Beijing), 2021, 43(3): 226-239.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Summary of the tool for estimating admixture time (LD-based)"
| 工具 | 交流模型 | 相关链接 | 参考文献 |
|---|---|---|---|
| ROLLOFF | HI (hybrid isolation) | | [76] |
| ALDER | HI | | [77] |
| MALDER | HI | | [77] |
| CAMer | HI, GA (gradual admixture), CGF (continuous gene flow), GA-I (GA-Isolation), CGF-I (CGF-Isolation) | | [78] |
| iMAAPs | HI, GA, CGF, GA-I, CGF-I | | [79] |
Table 2
Summary of the tool for estimating admixture time (Haplotype/ancestry block size distribution-based)"
| 工具 | 交流模型 | 相关链接 | 参考文献 |
|---|---|---|---|
| StepPCO | HI | | [72] |
| adwave | HI, Dual-admixture | | [71] |
| HAPMIX | HI | | [74] |
| MultiWaveInfer | HI, GA, CGF | | [80] |
| GLOBETROTTER | HI, GA, CGF | | [81] |
| tracts | HI, CGF | | [82] |
| Ancestry_HMM | HI | | [83] |
| [1] | PanZY, HeXY, WangXY, GuoXF, CaoXH, HuWP, DiR, LiuQY, ChuMX. Selection signature in domesticated animals. Hereditas(Beijing), 2016, 38(12): 1069- 1080. |
| 潘章源, 贺小云, 王翔宇, 郭晓飞, 曹晓涵, 胡文萍, 狄冉, 刘秋月, 储明星. 家养动物选择信号研究进展. 遗传, 2016, 38(12): 1069- 1080. | |
| [2] | ZederMA. Core questions in domestication research. Proc Natl Acad Sci USA, 2015, 112(11): 3191- 3198. |
| [3] | GambleC, DaviesW, PettittP, RichardsM. Climate change and evolving human diversity in Europe during the Last Glacial. Philos Trans R Soc Lond B Biol Sci, 2004, 359( 1442): 243- 254. |
| [4] | FabriceT. Animal domestication: A brief overview. London: IntechOpen. 2019. |
| [5] | ZederMA. The domestication of animals. J Anthropol Res, 1982,9(4):321- 327. |
| [6] | LarsonG, PipernoDR, AllabyRG, PuruggananMD, AnderssonL, Arroyo-KalinM, BartonL, ClimerVigueira C, DenhamT, DobneyK, DoustAN, GeptsP, GilbertMTP, GremillionKJ, LucasL, LukensL, MarshallFB, OlsenKM, PiresJC, RichersonPJ, deCasas RR, SanjurOI, ThomasMG, FullerDQ. Current perspectives and the future of domestication studies. Proc Natl Acad Sci USA, 2014, 111(17): 6139- 6146. |
| [7] | LiJ, ZhangYP. Advances in research of the origin and domestication of domestic animals. Biodiv Sci, 2009, 17(4): 1- 11. |
| 李晶, 张亚平. 家养动物的起源与驯化研究进展. 生物多样性, 2009, 17(4): 1- 11. | |
| [8] | DiamondJ. Evolution, consequences and future of plant and animal domestication. Nature, 2002,418(6898):700- 707. |
| [9] | EvershedRP, PayneS, SherrattAG, CopleyMS, CoolidgeJ, Urem-KotsuD, KotsakisK, OzdoğanM, OzdoğanAE, NieuwenhuyseO, AkkermansPMMG, BaileyD, AndeescuRR, CampbellS, FaridS, HodderI, YalmanN, OzbaşaranM, BiçakciE, GarfinkelY, LevyT, BurtonMM. Earliest date for milk use in the Near East and southeastern Europe linked to cattle herding. Nature, 2008, 455(7212): 528- 531. |
| [10] | OutramAK, StearNA, BendreyR, OlsenS, KasparovA, ZaibertV, ThorpeN, EvershedRP. The earliest horse harnessing and milking. Science, 2009,323(5919):1332- 1335. |
| [11] | DanielLH, AndrewGC. Principles of population genetics. 4th ed. Sinauer Associates, Sunderland, MA, 2006. |
| [12] | ShiY, LiHP. Population genomics: from classical statistics to supervised learning. Sci Sin Vitae, 2019, 49(4): 445- 455. |
| 施怿, 李海鹏. 群体基因组学方法:从经典统计学到有监督学习. 中国科学: 生命科学, 2019, 49(4): 445- 455. | |
| [13] | ZhengZQ. Population structure and genetic introgression from wild relatives in worldwide goat populations [Dissertation]. Northwest A&F University, 2019. |
| 郑竹清. 世界山羊群体遗传结构及其野生近缘种基因渗入研究[学位论文]. 西北农林科技大学, 2019. | |
| [14] | WangGD, ZhaiWW, YangHC, WangL, ZhongL, LiuYH, FanRX, YinTT, ZhuCL, PoyarkovAD, IrwinDM, HytönenMK, LohiH, WuCI, SavolainenP, ZhangYP. Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res, 2016, 26(1): 21- 33. |
| [15] | PetersJ, LebrasseurO, DengH, LarsonG. Holocene cultural history of Red jungle fowl ( Gallus gallus ) and its domestic descendant in East Asia . Quaternary Sci Rev, 2016,142:102- 119. |
| [16] | WangMS, ThakurM, PengMS, JiangY, FrantzLAF, LiM, ZhangJJ, WangS, PetersJ, OteckoNO, SuwannapoomC, GuoX, ZhengZQ, EsmailizadehA, HirimuthugodaNY, AshariH, SuladariS, ZeinMSA, KuszaS, SohrabiS, Kharrati-KoopaeeH, ShenQK, ZengL, YangMM, WuYJ, YangXY, LuXM, JiaXZ, NieQH, LamontSJ, LasagnaE, CeccobelliS, GunwardanaHGTN, SenasigeTM, FengSH, SiJF, ZhangH, JinJQ, LiML, LiuYH, ChenHM, MaC, DaiSS, BhuiyanAKFH, KhanMS, SilvaGLLP, LeTT, MwaiOA, IbrahimMNM, SuppleM, ShapiroB, HanotteO, ZhangGJ, LarsonG, HanJL, WuDD, ZhangYP. 863 genomes reveal the origin and domestication of chicken. Cell Res, 2020, 30(8):693- 701. |
| [17] | GaoF, LiHP. Application of computer simulators in population genetics. Hereditas(Beijing), 2016, 38(8): 707- 717. |
| 高峰, 李海鹏. 群体遗传学模拟软件应用现状. 遗传, 2016, 38(8): 707- 717. | |
| [18] | LiH, DurbinR. Inference of human population history from individual whole-genome sequences. Nature, 2011, 475(7357): 493- 496. |
| [19] | SchiffelsS, DurbinR. Inferring human population size and separation history from multiple genome sequences. Nat Genet, 2014,46(8):919- 925. |
| [20] | TerhorstJ, KammJA, SongYS. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet, 2017, 49(2): 303- 309. |
| [21] | QanbariS, StromTM, HabererG, WeigendS, GheyasAA, TurnerF, BurtDW, PreisingerR, GianolaD, SimianerH. A high resolution genome-wide scan for significant selective sweeps: an application to pooled sequence data in laying chickens. PLoS One, 2012, 7( 11): e49525. |
| [22] | LiHP, StephanW. Inferring the demographic history and rate of adaptive substitution in Drosophila. PLoS Genet, 2006, 2( 10): e166. |
| [23] | WakeleyJ, HeyJ. Estimating ancestral population parameters. Genetics, 1997,145(3):847- 855. |
| [24] | GutenkunstRN, HernandezRD, WilliamsonSH, BustamanteCD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet, 2009, 5( 10): e1000695. |
| [25] | ExcoffierL, DupanloupI, Huerta-SánchezE, SousaVC, FollM. Robust demographic inference from genomic and SNP data. PLoS Genet, 2013, 9( 10): e1003905. |
| [26] | BarbatoM, Orozco-terWengelP, TapioM, BrufordMW. Snep: a tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front Genet, 2015,6:109. |
| [27] | WangJL. Estimation of effective population sizes from data on genetic markers. Philos Trans R Soc Lond B Biol Sci, 2005,360( 1459):1395- 1409. |
| [28] | HillWG. Estimation of effective population size from data on linkage disequilibrium. Genet Res, 1981,38(3):209- 216. |
| [29] | CorbinLJ, LiuAYH, BishopSC, WoolliamsJA. Estimation of historical effective population size using linkage disequilibria with marker data. J Anim Breed Genet, 2012,129(4):257- 270. |
| [30] | SunnåkerM, BusettoAG, NumminenE, CoranderJ, FollM, DessimozC. Approximate Bayesian computation. PLoS Comput Biol, 2013, 9( 1): e1002803. |
| [31] | WegmannD, LeuenbergerC, NeuenschwanderS, ExcoffierL. Abctoolbox: A versatile toolkit for approximate Bayesian computations. BMC Bioinformatics, 2010,11(116). |
| [32] | SanchezT, CuryJ, CharpiatG, JayF. Deep learning for population size history inference: design, comparison and combination with approximate Bayesian computation. Mol Ecol Resour, 2020. |
| [33] | Tiago Do Prado Paim, PatríciaI, SamuelRP, AlexandreRC, Concepta Margaret Mcmanus Pimentel. Detection and evaluation of selection signatures in sheep. Pesqui Agropecu Bras, 2018,53(5):527- 539. |
| [34] | XueZYY, SongXW, WuLH, WangLZ, CuiJA, SunZJ, ZhangZ, MaYL. The identification methods of selection signatures in livestock and its statistical problems. Acta Vet Et Zootech Sin, 2018, 49(6): 1099- 1107. |
| 薛周舣源, 宋显威, 吴林慧, 王露珍, 崔家安, 孙章健, 张政, 马云龙. 畜禽选择信号检测方法及其统计学问题. 畜牧兽医学报, 2018, 49(6): 1099- 1107. | |
| [35] | EisenhaberF. Discovering biomolecular mechanisms with computational biology. Springer, Boston, MA, 2006. |
| [36] | PenningsPS, HermissonJ. Soft sweeps II—molecular population genetics of adaptation from recurrent mutation or migration. Mol Biol Evol, 2006,23(5):1076- 1084. |
| [37] | HermissonJ, PenningsPS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics, 2005,169(4):2335- 2352. |
| [38] | SuzukiY. Statistical methods for detecting natural selection from genomic data. Genes Genet Syst, 2010,85(6):359- 376. |
| [39] | NielsenR. Molecular signatures of natural selection. Annu Rev Genet, 2005,39:197- 218. |
| [40] | LohmuellerKE, BustamanteCD, ClarkAG. Detecting directional selection in the presence of recent admixture in African-Americans. Genetics, 2011,187(3):823- 835. |
| [41] | WangYZ, ZhaoYQ. Research progress of genomic signature of selection and its detection methods. Acta Ecol Anim Domas, 2019, 40(5): 1- 6. |
| 王宇占, 赵毅强. 基因组水平的选择信号及其检测方法研究进展. 家畜生态学报, 2019, 40(5): 1- 6. | |
| [42] | de Simoni Gouveia JJ, daSilva MVGB, PaivaSR, de Oliveira SMP. Identification of selection signatures in livestock species. Genet Mol Biol, 2014, 37(2): 330- 342. |
| [43] | ChenH, PattersonN, ReichD. Population differentiation as a test for selective sweeps. Genome Res, 2010,20(3):393- 402. |
| [44] | OleksykTK, SmithMW, O'BrienSJ. Genome-wide scans for footprints of natural selection. Philos Trans R Soc Lond B Biol Sci, 2010, 365( 1537): 185- 205. |
| [45] | GrossmanSR, ShlyakhterI, KarlssonEK, ByrneEH, MoralesS, FriedenG, HostetterE, AngelinoE, GarberM, ZukO, LanderES, SchaffnerSF, SabetiPC. A composite of multiple signals distinguishes causal variants in regions of positive selection. Science, 2010, 327(5967): 883- 886. |
| [46] | UtsunomiyaYT, PérezO'Brien AM, SonstegardTS, VanTassell CP, doCarmo AS, MészárosG, SölknerJ, GarciaJF. Detecting loci under recent positive selection in dairy and beef cattle by combining different genome- wide scan methods. PLoS One, 2013,8(5): e64280. |
| [47] | RandhawaIAS, KhatkarMS, ThomsonPC, RaadsmaHW. Composite selection signals can localize the trait specific genomic regions in multi-breed populations of cattle and sheep. BMC Genet, 2014,15:34. |
| [48] | MaY, DingX, QanbariS, WeigendS, ZhangQ, SimianerH. Properties of different selection signature statistics and a new strategy for combining them. Heredity(Edinb), 2015,115(5):426- 436. |
| [49] | BiswasS, AkeyJM. Genomic insights into positive selection. Trends Genet, 2006,22(8):437- 446. |
| [50] | SchriderDR, KernAD. Supervised machine learning for population genetics: a new paradigm. Trends Genet, 2018,34(4):301- 312. |
| [51] | KotsiantisSB. Supervised machine learning: a review of classification techniques. Informatica (lith Acad Sci), 2007, 31: 3- 24. |
| [52] | LinK, LiHP, SchlöttererC, FutschikA. Distinguishing positive selection from neutral evolution: boosting the performance of summary statistics. Genetics, 2011, 187(1): 229- 244. |
| [53] | LiangZX. Analysis of pig domestication and ancestry using genomewide SNP information[Dissertation]. China Agricultural University, 2019. |
| 梁作翔. 利用全基因组SNP信息研究家猪的驯化及祖先来源[学位论文]. 中国农业大学, 2019. | |
| [54] | MalaspinasAS, MalaspinasO, EvansSN, SlatkinM. Estimating allele age and selection coefficient from time-serial data. Genetics, 2012, 192(2): 599- 607. |
| [55] | PrzeworskiM. Estimating the time since the fixation of a beneficial allele. Genetics, 2003,164(4):1667- 1676. |
| [56] | LaurentS, PfeiferSP, SettlesML, HunterSS, HardwickKM, OrmondL, SousaVC, JensenJD, RosenblumEB. The population genomics of rapid adaptation: disentangling signatures of selection and demography in white sands lizards. Mol Ecol, 2016, 25(1): 306- 323. |
| [57] | MedinaP, ThornlowB, NielsenR, Corbett-DetigR. Estimating the timing of multiple admixture pulses during local ancestry inference. Genetics, 2018,210(3):1089- 1107. |
| [58] | ChengCH, HuangY. Construction and application of phylogenetic network. Entomotaxonomia, 2008, 30(3): 215- 221. |
| 程春花, 黄原. 系统发育网络的构建与应用. 昆虫分类学报, 2008, 30(3): 215- 221. | |
| [59] | DobneyK, LarsonG. Genetics and animal domestication: New windows on an elusive process. J Zool, 2006, 269(2): 261- 271. |
| [60] | MarshallFB, DobneyK, DenhamT, CaprilesJM. Evaluating the roles of directed breeding and gene flow in animal domestication. Proc Natl Acad Sci USA, 2014, 111(17): 6153- 6158. |
| [61] | WinklerCA, NelsonGW, SmithMW. Smith. Admixture mapping comes of age. Annu Rev Genomics Hum Genet, 2010,11:65- 89. |
| [62] | RacimoF, SankararamanS, NielsenR, Huerta-SánchezE. Evidence for archaic adaptive introgression in humans. Nat Rev Genet, 2015,16(6):359- 371. |
| [63] | AiHS, FangXD, YangB, HuangZY, ChenH, MaoLK, ZhangF, ZhangL, CuiLL, HeWM, YangJ, YaoXM, ZhouLS, HanLJ, LiJ, SunSL, XieXH, LaiBX, SuY, LuY, YangH, HuangT, DengWJ, NielsenR, RenJ, HuangLS. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet, 2015,47(3):217- 225. |
| [64] | LiangSY, ZhouZK, HouSS. The research progress of farm animal genomics based on sequencing technologies. Hereditas(Beijing), 2017, 39(4): 276- 292. |
| 梁素芸, 周正奎, 侯水生. 基于测序技术的畜禽基因组学研究进展. 遗传, 2017, 39(4): 276- 292. | |
| [65] | PattersonN, MoorjaniP, LuoY, MallickS, RohlandN, ZhanYP, GenschoreckT, WebsterT, ReichD. Ancient admixture in human history. Genetics, 2012,192(3):1065- 1093. |
| [66] | ReichD, ThangarajK, PattersonN, PriceAL, SinghL. Reconstructing Indian population history. Nature, 2009,461(7263):489- 494. |
| [67] | GreenRE, KrauseJ, BriggsAW, MaricicT, StenzelU, KircherM, PattersonN, LiH, ZhaiWW, FritzMHY, HansenNF, DurandEY, MalaspinasAS, JensenJD, Marques-BonetT, AlkanC, PrüferK, MeyerM, BurbanoHA, GoodJM, SchultzR, Aximu-PetriA, ButthofA, HöberB, HöffnerB, SiegemundM, WeihmannA, NusbaumC, LanderES, RussC, NovodN, AffourtitJ, EgholmM, VernaC, RudanP, BrajkovicD, KucanŽ, GušicI, DoronichevVB, GolovanovaLV, Lalueza-FoxC, dela Rasilla M, ForteaJ, RosasA, SchmitzRW, JohnsonPLF, EichlerEE, FalushD, BirneyE, MullikinJC, SlatkinM, NielsenR, KelsoJ, LachmannM, ReichD, PääboS. A draft sequence of the Neandertal genome. Science, 2010,328(5979):710- 722. |
| [68] | PeaseJB, HahnMW. Detection and polarization of introgression in a five-taxon phylogeny. Syst Biol, 2015,64(4):651- 662. |
| [69] | ZhengYC, JankeA. Gene flow analysis method, the D -statistic, is robust in a wide parameter space . BMC Bioinformatics, 2018, 19( 1): 10. |
| [70] | FrantzLAF, SchraiberJG, MadsenO, MegensHJ, BosseM, PaudelY, SemiadiG, MeijaardE, LiN, CrooijmansRPMA, ArchibaldAL, SlatkinM, SchookLB, LarsonG, GroenenMAM. Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol, 2013,14(9): R107. |
| [71] | SandersonJ, SudoyoH, KarafetTM, HammerMF, CoxMP. Reconstructing past admixture processes from local genomic ancestry using wavelet transformation. Genetics, 2015, 200(2): 469- 481. |
| [72] | PugachI, MatveyevR, WollsteinA, KayserM, StonekingM. Dating the age of admixture via wavelet transform analysis of genome-wide data . Genome Biol, 2011, 12( 2): R19. |
| [73] | XuSH, HuangW, QianJ, JinL. Analysis of genomic admixture in Uyghur and its implication in mapping strategy. Am J Hum Genet, 2008,82(4):883- 894. |
| [74] | PriceAL, TandonA, PattersonN, BarnesKC, RafaelsN, RuczinskiI, BeatyTH, MathiasR, ReichD, MyersS. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet, 2009,5(6): e1000519. |
| [75] | ChimusaER, DefoJ, ThamiPK, AwanyD, MulisaDD, AllaliI, GhazalH, MoussaA, MazanduGK. Dating admixture events is unsolved problem in multi-way admixed populations. Brief Bioinform, 2020, 21(2): 144- 155. |
| [76] | MoorjaniP, PattersonN, HirschhornJN, KeinanA, HaoL, AtzmonG, BurnsE, OstrerH, PriceAL, ReichD. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet, 2011,7(4): e1001373. |
| [77] | LohPR, LipsonM, PattersonN, MoorjaniP, PickrellJK, ReichD, BergerB. Inferring admixture histories of human populations using linkage disequilibrium. Genetics, 2013,193(4):1233- 1254. |
| [78] | ZhouY, QiuHX, XuSH. Modeling continuous admixture using admixture-induced linkage disequilibrium. Sci Rep, 2017,7:43054. |
| [79] | ZhouY, YuanK, YuY, NiX, XieP, XingEP, XuS. Inference of multiple-wave population admixture by modeling decay of linkage disequilibrium with polynomial functions. Heredity (Edinb), 2017,118(5):503- 510. |
| [80] | NiXM, YuanK, YangX, FengQD, GuoW, MaZM, XuSH. Inference of multiple-wave admixtures by length distribution of ancestral tracks. Heredity (Edinb), 2018, 121(1): 52- 63. |
| [81] | HellenthalG, BusbyGBJ, BandG, WilsonJF, CapelliC, FalushD, MyersS. A genetic atlas of human admixture history. Science, 2014, 343(6172): 747- 751. |
| [82] | NiXM, YangX, GuoW, YuanK, ZhouY, MaZM, XuSH. Length distribution of ancestral tracks under a general admixture model and its applications in population history inference. Sci Rep, 2016,6:20048. |
| [83] | Corbett-DetigR, NielsenR. A hidden Markov model approach for simultaneously estimating local ancestry and admixture time using next generation sequence data in samples of arbitrary ploidy. PLoS Genet, 2017, 13( 1): e1006529. |
| [84] | LawsonDJ, HellenthalG, MyersS, FalushD. Inference of population structure using dense haplotype data. PLoS Genet, 2012, 8( 1): e1002453. |
| [85] | GalaverniM, CanigliaR, PaganiL, FabbriE, BoattiniA, RandiE. Disentangling timing of admixture, patterns of introgression, and phenotypic indicators in a hybridizing wolf population. Mol Biol Evol, 2017,34(9):2324- 2339. |
| [86] | ZhuYL, LiWB, YangB, ZhangZY, AiHS, RenJ, HuangLS. Signatures of selection and interspecies introgression in the genome of Chinese domestic pigs. Genome Biol Evol, 2017, 9(10): 2592- 2603. |
| [87] | ChenH, HuangM, YangB, WuZP, DengZ, HouY, RenJ, HuangLS. Introgression of Eastern Chinese and Southern Chinese haplotypes contributes to the improvement of fertility and immunity in European modern pigs. Gigascience, 2020,9(3): giaa014. |
| [88] | RubinCJ, ZodyMC, ErikssonJ, MeadowsJRS, SherwoodE, WebsterMT, JiangL, IngmanM, SharpeT, KaS, HallböökF, BesnierF, CarlborgO, Bed'homB, Tixier-BoichardM, JensenP, SiegelP, Lindblad-TohK, AnderssonL. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature, 2010,464(7288):587- 591. |
| [89] | LiDY, LiY, LiM, CheTD, TianSL, ChenBL, ZhouXM, ZhangGL, GaurU, LuoMJ, TianK, HeMN, HeS, XuZX, JinL, TangQZ, DaiYF, XuHL, HuYD, ZhaoXL, YinHD, WangY, ZhouRJ, YangCW, DuHR, JiangXS, ZhuQ, LiMZ. Population genomics identifies patterns of genetic diversity and selection in chicken. BMC Genomics, 2019, 20( 1): 263. |
| [90] | ZhangCY, LinD, WangYZ, PengDZ, LiHF, FeiJ, ChenKW, YangN, HuXX, ZhaoYQ, LiN. Widespread introgression in Chinese indigenous chicken breeds from commercial broiler. Evol Appl, 2019,12(3):610- 621. |
| [91] | ZederMA, HesseB. The initial domestication of goats ( Capra hircus ) in the Zagros Mountains 10, 000 years ago. Science, 2000, 287(5461): 2254- 2257. |
| [92] | WangFH, ZhangL, LiXK, FanYX, QiaoX, GongG, YanXC, ZhangLT, WangZY, WangRJ, LiuZH, WangZX, HeLB, ZhangYJ, LiJQ, ZhaoYH, SuR. Progress in goat genome studies. Hereditas(Beijing), 2019, 41(10): 928- 938. |
| 王凤红, 张磊, 李晓凯, 范一星, 乔贤, 龚高, 严晓春, 张令天, 王志英, 王瑞军, 刘志红, 王志新, 何利兵, 张燕军, 李金泉, 赵艳红, 苏蕊. 山羊基因组研究进展. 遗传, 2019, 41(10): 928- 938. | |
| [93] | LiXK, WangG, QiaoX, FanY, ZhangL, MaYH, NieRX, WangRJ, HeLB, SuR. Research progress on whole-genome sequencing on important domesticated animals. Biotechnol Bull, 2018, 34(6): 11- 21. |
| 李晓凯, 王贵, 乔贤, 范一星, 张磊, 马宇浩, 聂瑞雪, 王瑞军, 何利兵, 苏蕊. 全基因组测序在重要家畜上的研究进展. 生物技术通报, 2018, 34(6): 11- 21. | |
| [94] | CaoYH, XuSS, ShenM, ChenZH, GaoL, LvFH, XieXL, WangXH, YangH, LiuCB, ZhouP, WanPC, ZhangYS, YangJQ, PiWH, EerH, BerryDP, BarbatoM, EsmailizadehA, NosratiM, Salehian-DehkordiH, Dehghani-QanatqestaniM, DotsevAV, DeniskovaTE, ZinovievaNA, BremG, ŠtěpánekO, CianiE, WeimannC, ErhardtG, MwacharoJM, AhbaraA, HanJL, HanotteO, MillerJM, SimZ, ColtmanD, KantanenJ, BrufordMW, LenstraJA, KijasJ, LiMH. Historical introgression from wild relatives enhanced climatic adaptation and resistance to pneumonia in sheep. Mol Biol Evol, 2020,17:msaa236. |
| [95] | ZhengZQ, WangXH, LiM, LiYJ, YangZR, WangXL, PanXY, GongM, ZhangY, GuoYW, WangY, LiuJ, CaiYD, ChenQM, OkpekuM, ColliL, CaiDW, WangK, HuangSS, SonstegardTS, EsmailizadehA, ZhangWG, ZhangTT, XuYB, XuNY, YangY, HanJL, ChenL, LesurJ, DalyKG, BradleyDG, HellerR, ZhangGJ, WangW, ChenYL, JiangY. The origin of domestication genes in goats. Sci Adv, 2020, 6( 21): eaaz5216. |
| [96] | TerhorstJ, KammJA, SongYS. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet, 2017, 49(2): 303- 309. |
| [97] | OrmondL, FollM, EwingGB, PfeiferSP, JensenJD. Inferring the age of a fixed beneficial allele. Mol Ecol, 2016, 25(1): 157- 169. |
| [98] | FrantzLA, SchraiberJG, MadsenO, MegensHJ, CaganA, BosseM, PaudelY, CrooijmansRPMA, LarsonG, GroenenMAM. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet, 2015, 47(10): 1141- 1148. |
| [99] | FreudenthalJA, AnkenbrandMJ, GrimmDG, KorteA. GWAS-Flow: A GPU accelerated framework for efficient permutation based genome-wide association studies. BioRxiv, 2019,1:783100. |
| [100] | BuLN, WangQ, GuWJ, YangRF, ZhuD, SongZ, LiuXJ, ZhaoYQ. Improving read alignment through the generation of alternative reference via iterative strategy . Sci Rep, 2020, 10( 1): 18712. |
| [1] | Luyan Tian, Xiaozhen Huang. Application value of protein phase separation mechanism of flowering regulation in de novo domestication [J]. Hereditas(Beijing), 2023, 45(9): 754-764. |
| [2] | Xiaoping Lian, Guangfu Huang, Yujiao Zhang, Jing Zhang, Fengyi Hu, Shilai Zhang. The discovery and utilization of favorable genes in Oryza longistaminata [J]. Hereditas(Beijing), 2023, 45(9): 765-780. |
| [3] | Liumei Jian, Yingjie Xiao, Jianbing Yan. De novo domestication: a new way for crop design and breeding [J]. Hereditas(Beijing), 2023, 45(9): 741-753. |
| [4] | Biwei Lai, Lei Chen, Sijia Lu. The current status of photoperiod adaptability in soybean [J]. Hereditas(Beijing), 2023, 45(9): 793-800. |
| [5] | Fei Wang, Meng Wang, Xinghua Zhang, Keli Yu, Lianbin Zheng, Yajun Yang. Genetic substructure analysis of three isolated populations in southwest China [J]. Hereditas(Beijing), 2022, 44(5): 424-431. |
| [6] | Meng Fu, Yan Li. The origin and domestication history of domestic horses and the domestication characteristics of breeds [J]. Hereditas(Beijing), 2022, 44(3): 216-229. |
| [7] | Jinyan Yang, Xueqin Liu, Tianqi Wen, Yuhong Sun, Ying Yu. Progress on lncRNA regulated disease resistance traits in domesticated animals [J]. Hereditas(Beijing), 2021, 43(7): 654-664. |
| [8] | Xiaojuan Wang, Enfang Qian, Yue Li, Zhengyang Song, Hui Zhao, Hexin Xie, Caixia Li, Jiang Huang, Li Jiang. A genetic sub-structure study of the Tibetan population in Southwest China [J]. Hereditas(Beijing), 2020, 42(6): 565-576. |
| [9] | Xinping Yang,Yuan Yu,Cao Xu. De novo domestication to create new crops [J]. Hereditas(Beijing), 2019, 41(9): 827-835. |
| [10] | Chun Lin,Zhengjie Liu,Yumei Dong,Michel Vales,Zichao Mao. Domesticated cultivation and genetic breeding of Chenopodium quinoa [J]. Hereditas(Beijing), 2019, 41(11): 1009-1022. |
| [11] | Jian Hu,Yiren Zhou,Jialin Ding,Zhiyuan Wang,Ling Liu,Yekai Wang,Huiling Lou,Shouyi Qiao,Yanhua Wu. Simplification of genotyping techniques of the ABO blood type experiment and exploration of population genetics [J]. Hereditas(Beijing), 2017, 39(5): 423-429. |
| [12] | Feng Gao, Haipeng Li. Application of computer simulators in population genetics [J]. Hereditas(Beijing), 2016, 38(8): 707-717. |
| [13] | Yunsheng Wang. Research progress of plant population genomics based on high-throughput sequencing [J]. Hereditas(Beijing), 2016, 38(8): 688-699. |
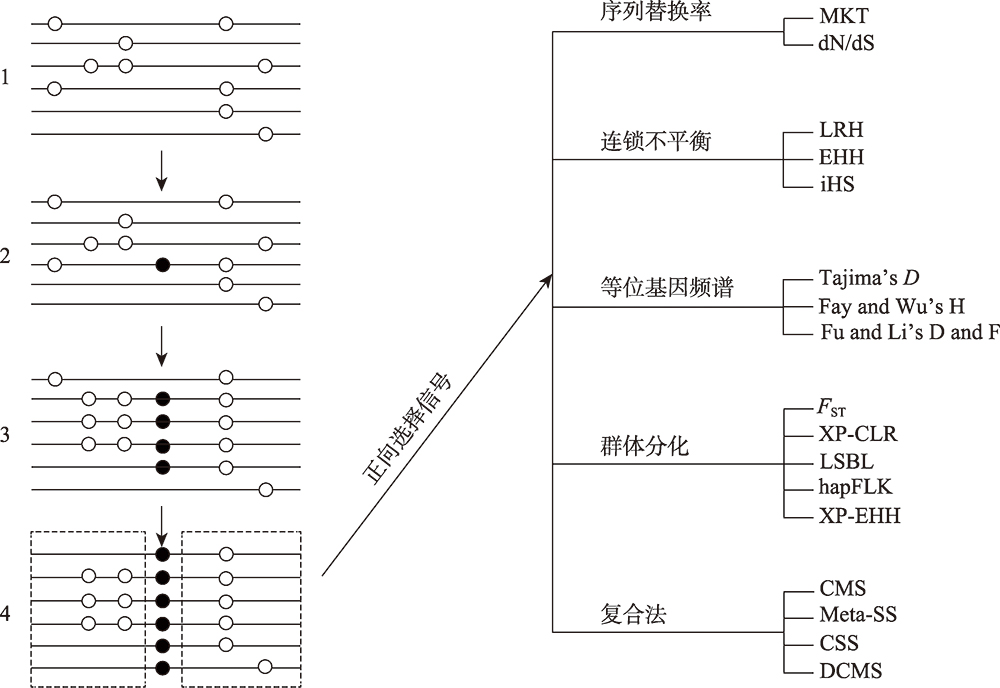
| [14] | Zhangyuan Pan, Xiaoyun He, Xiangyu Wang, Xiaofei Guo, Xiaohan Cao, Wenping Hu, Ran Di, Qiuyue Liu, Mingxing Chu. Selection signature in domesticated animals [J]. Hereditas(Beijing), 2016, 38(12): 1069-1080. |
| [15] | Chengcai Chu. The Cover story description [J]. HEREDITAS, 2012, 34(11): 1389-1389. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||