Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (8): 775-791.doi: 10.16288/j.yczz.21-132
• Orginal Articles • Previous Articles Next Articles
Serial deletions of tandem reverse CTCF sites reveal balanced HOXD regulatory landscape of enhancers
Ling Wang( ), Jinhuan Li, Haiyan Huang(
), Jinhuan Li, Haiyan Huang( ), Qiang Wu(
), Qiang Wu( )
)
- Center for Comparative Biomedicine, Key laboratory of Systems Biomedicine (Ministry of Education), Institute of Systems Biomedicine, Shanghai Jiao Tong University, Shanghai 200240, China
-
Received:2021-04-11Revised:2021-05-12Online:2021-08-20Published:2021-06-30 -
Contact:Huang Haiyan,Wu Qiang E-mail:wanglingmail0613@163.com;hy_huang@sjtu.edu.cn;qwu123@gmail.com -
Supported by:Supported by the National Natural Science Foundation of China Nos(31800636);Supported by the National Natural Science Foundation of China Nos(31630039);Supported by the National Natural Science Foundation of China Nos(91940303);Science and Technology Commission of Shanghai Municipality No(19JC1412500)
Cite this article
Ling Wang, Jinhuan Li, Haiyan Huang, Qiang Wu. Serial deletions of tandem reverse CTCF sites reveal balanced HOXD regulatory landscape of enhancers[J]. Hereditas(Beijing), 2021, 43(8): 775-791.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
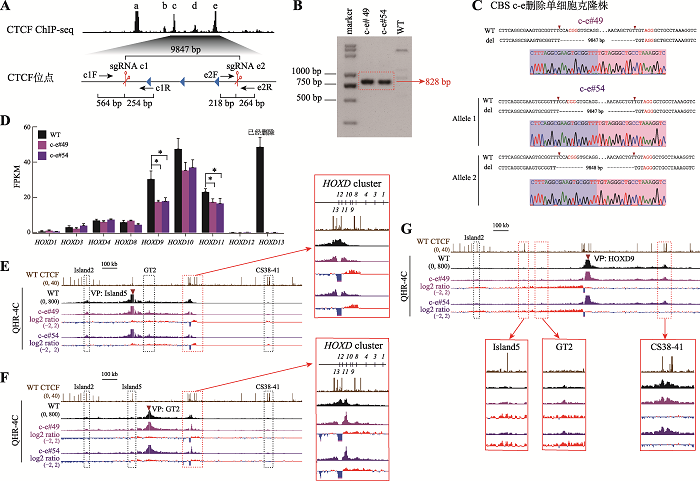
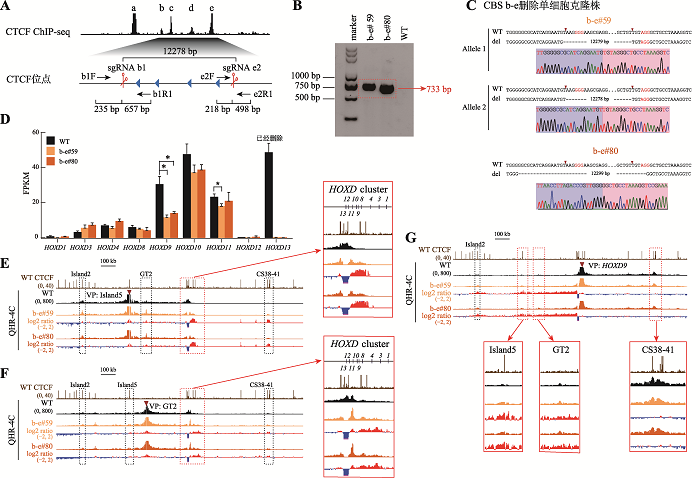
Table 1
Primers used in this study"
| 类型 | 引物名称 | 序列(5′→3′) |
|---|---|---|
| PCR | a1F | TTCCAGCACCTCGGCTTTGTC |
| a1R | CCCACTTTCCACCTCTGTCCTG | |
| b1F | GTCCGCCCGTGAGCTTCTGAA | |
| b1R1 | CTCACAGCAGCCGAAACCG | |
| c1F | TGATGCAGCCTCTGTGACCG | |
| c1R | AGTTTTCCCGTGGCGTCTGA | |
| e1F | TTCCCTGTCCCAGCTTGATTTC | |
| e1R | TCAACAGTGAAGGGCGGTGC | |
| e2F | CAAGCCACTCTCCCGCCACTA | |
| e2R | TCGCTCTCGTCCTCTCTTGGG | |
| e2R1 | GGCTCCTGCACTGAGACCACA | |
| sgRNA | sgRNA a1F | ACCGCGAAGAGTGCGGGAGAACGG |
| sgRNA a1R | AAACCCGTTCTCCCGCACTCTTCG | |
| sgRNA b1F | ACCGGGCGCATCAGGAATGTAAG | |
| sgRNA b1R | AAACCTTACATTCCTGATGCGCC | |
| sgRNA c1F | ACCGCAGGCGAAGTGCGGTTTCCA | |
| sgRNA c1R | AAACTGGAAACCGCACTTCGCCTG | |
| sgRNA e1F | ACCGAACTGTGCTCAAACGCTCTC | |
| sgRNA e1R | AAACGAGAGCGTTTGAGCACAGTT | |
| sgRNA e2F | ACCGGAGGCGCAAACAGCTGTTGT | |
| sgRNA e2R | AAACACAACAGCTGTTTGCGCCTC | |
| Index | Island5-bioprimer | 5′biotin-AAACACAAATGCATCAACCTG |
| GT2-bioprimer | 5′biotin-GAGCCAAACTGTACCCCTAGC | |
| HOXD9-bioprimer | 5′biotin-ACCGACTAGTTCGCAGGCT | |
| Island5-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATGCATCTCATGAAGCTGGCATCT | |
| GT2-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGTAACTTTAGCTAAACCAAGGCCT | |
| HOXD9-P5 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCTGCAGCCTCCACCATTG | |
| P7-index-1 | CAAGCAGAAGACGGCATACGAGATCGAGTAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-2 | CAAGCAGAAGACGGCATACGAGATTCTCCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-3 | CAAGCAGAAGACGGCATACGAGATAATGAGCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-4 | CAAGCAGAAGACGGCATACGAGATGGAATCTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-5 | CAAGCAGAAGACGGCATACGAGATTTCTGAATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-6 | CAAGCAGAAGACGGCATACGAGATACGAATTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-7 | CAAGCAGAAGACGGCATACGAGATAGCTTCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-8 | CAAGCAGAAGACGGCATACGAGATGCGCATTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-9 | CAAGCAGAAGACGGCATACGAGATCATAGCCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-10 | CAAGCAGAAGACGGCATACGAGATTTCGCGGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-11 | CAAGCAGAAGACGGCATACGAGATGCGCGAGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-12 | CAAGCAGAAGACGGCATACGAGATCTATCGCTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-13 | CAAGCAGAAGACGGCATACGAGATAGAGTACTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-14 | CAAGCAGAAGACGGCATACGAGATGCTCCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-15 | CAAGCAGAAGACGGCATACGAGATCATGAGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-16 | CAAGCAGAAGACGGCATACGAGATTGAATCGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-17 | CAAGCAGAAGACGGCATACGAGATGTCTGACTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-18 | CAAGCAGAAGACGGCATACGAGATCTGAATGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-19 | CAAGCAGAAGACGGCATACGAGATCGCTTCTGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-20 | CAAGCAGAAGACGGCATACGAGATTCGCATGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-21 | CAAGCAGAAGACGGCATACGAGATAATAGCAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-22 | CAAGCAGAAGACGGCATACGAGATGTCGCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-23 | CAAGCAGAAGACGGCATACGAGATACGCGATAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-24 | CAAGCAGAAGACGGCATACGAGATTGATCGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-25 | CAAGCAGAAGACGGCATACGAGATCCGCATGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-26 | CAAGCAGAAGACGGCATACGAGATCCACAATCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-27 | CAAGCAGAAGACGGCATACGAGATGATGTTCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT |
| [1] |
Wu Q, Liu PF, Wang LY. Many facades of CTCF unified by its coding for three-dimensional genome architecture. J Genet Genomics, 2020, 47(8):407-424.
doi: 10.1016/j.jgg.2020.06.008 |
| [2] |
Guo Y, Monahan K, Wu HY, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci USA, 2012, 109(51):21081-21086.
doi: 10.1073/pnas.1219280110 |
| [3] |
Guo Y, Xu Q, Canzio D, Shou J, Li JH, Gorkin DU, Jung I, Wu HY, Zhai YN, Tang YX, Lu YC, Wu YH, Jia ZL, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015, 162(4):900-910.
doi: 10.1016/j.cell.2015.07.038 pmid: 26276636 |
| [4] | Zhai YN, Xu Q, Guo Y, Wu Q. Characterization of a cluster of CTCF-binding sites in a protocadherin regulatory region. Hereditas(Beijing), 2016, 38(4):323-336. |
| 翟亚男, 许泉, 郭亚, 吴强. 原钙粘蛋白基因簇调控区域中成簇的CTCF结合位点分析. 遗传, 2016, 38(4):323-336. | |
| [5] |
Yin M, Wang J, Wang M, Li X, Zhang M, Wu Q, Wang Y. Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res, 2017, 27(11):1365-1377.
doi: 10.1038/cr.2017.131 |
| [6] | Guo Y, Wu Q. Inversion of CTCF binding sites by DNA fragment editing alters genome topology and enhancer/ promoter functions. Hereditas(Beijing), 2015, 37(10):1073-1074. |
| 郭亚, 吴强. 采用DNA片段编辑技术反转CTCF结合位点改变基因组拓扑结构和增强子与启动子功能. 遗传, 2015, 37(10):1073-1074. | |
| [7] |
Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol, 1996, 16(6):2802-2813.
pmid: 8649389 |
| [8] |
Chen HB, Tian Y, Shu WJ, Bo XC, Wang SQ. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS One, 2012, 7(7):e41374.
doi: 10.1371/journal.pone.0041374 |
| [9] |
Nasmyth K. Disseminating the genome: Joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet, 2001, 35:673-745.
pmid: 11700297 |
| [10] |
Kim Y, Shi ZB, Zhang HS, Finkelstein IJ, Yu HT. Human cohesin compacts DNA by loop extrusion. Science, 2019, 366(6471):1345-1349.
doi: 10.1126/science.aaz4475 |
| [11] |
Lu YJ, Shou J, Jia ZL, Wu YH, Li JH, Guo Y, Wu Q. Genetic evidence for asymmetric blocking of higher-order chromatin structure by CTCF/cohesin. Protein Cell, 2019, 10(12):914-920.
doi: 10.1007/s13238-019-00656-y |
| [12] | Zheng XF, Huang HY, Wu Q. Chromatin architectural protein CTCF regulates gene expression of the UGT1 cluster. Hereditas(Beijing), 2019, 41(6):509-523. |
| 郑晓飞, 黄海燕, 吴强. 染色质架构蛋白CTCF调控UGT1基因簇的表达. 遗传, 2019, 41(6):509-523. | |
| [13] |
Jia ZL, Li JW, Ge X, Wu YH, Guo Y, Wu Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol, 2020, 21(1):75.
doi: 10.1186/s13059-020-01984-7 |
| [14] | Wu YH, Jia ZL, Ge X, Wu Q. Three-dimensional genome architectural CCCTC-binding factor makes choice in duplicated enhancers at Pcdhα locus. Sci China Life Sci, 2020, 63(6):835-844. |
| [15] |
Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell, 2016, 164(6):1110-1121.
doi: S0092-8674(16)30073-3 pmid: 26967279 |
| [16] |
Nichols MH, Corces VG. A tethered-inchworm model of SMC DNA translocation. Nat Struct Mol Biol, 2018, 25(10):906-910.
doi: 10.1038/s41594-018-0135-4 |
| [17] |
Wu Q, Jia ZL. Wiring the brain by clustered protocadherin neural codes. Neurosci Bull, 2021, 37(1):117-131.
doi: 10.1007/s12264-020-00578-4 |
| [18] | Lin SG, Ba ZQ, Alt FW, Zhang Y. RAG chromatin scanning during V(D)J recombination and chromatin loop extrusion are related processes. Adv Immunol, 2018, 139:93-135. |
| [19] |
Chen L, Carico Z, Shih HY, Krangel MS. A discrete chromatin loop in the mouse Tcra-Tcrd locus shapes the TCRdelta and TCRalpha repertoires. Nat Immunol, 2015, 16(10):1085-1093.
doi: 10.1038/ni.3232 |
| [20] | Majumder K, Koues OI, Chan EAW, Kyle KE, Horowitz JE, Yang-Iott K, Bassing CH, Taniuchi I, Krangel MS, Oltz EM. Lineage-specific compaction of Tcrb requires a chromatin barrier to protect the function of a long-range tethering element. J Exp Med, 2015, 212(1):107-120. |
| [21] | Rodríguez-Carballo E, Lopez-Delisle L, Zhan Y, Fabre PJ, Beccari L, El-Idrissi I, Huynh THN, Ozadam H, Dekker J, Duboule D. The HoxD cluster is a dynamic and resilient TAD boundary controlling the segregation of antagonistic regulatory landscapes. Genes Dev, 2017, 31(22):2264-2281. |
| [22] | Rodríguez-Carballo E, Lopez-Delisle L, Yakushiji- Kaminatsui N, Ullate-Agote A, Duboule D. Impact of genome architecture on the functional activation and repression of Hox regulatory landscapes. BMC Biol, 2019, 17(1):55. |
| [23] | Rodríguez-Carballo E, Lopez-Delisle L, Willemin A, Beccari L, Gitto S, Mascrez B, Duboule D. Chromatin topology and the timing of enhancer function at the HoxD locus. Proc Natl Acad Sci USA, 2020, 117(49):31231-31241. |
| [24] |
Jia ZL, Wu Q. Clustered protocadherins emerge as novel susceptibility loci for mental disorders. Front Neurosci, 2020, 14:587819.
doi: 10.3389/fnins.2020.587819 |
| [25] |
Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci USA, 2012, 109(43):17507-17512.
doi: 10.1073/pnas.1111941109 |
| [26] |
Lewis EB. A gene complex controlling segmentation in Drosophila. Nature, 1978, 276(5688):565-570.
doi: 10.1038/276565a0 |
| [27] | Mallo M. Reassessing the role of Hox genes during vertebrate development and evolution. Trends Genet, 2018, 34(3):209-217. |
| [28] |
Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science, 2003, 301(5631):331-333.
doi: 10.1126/science.1085753 |
| [29] | Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science, 2013, 340(6137):1234167. |
| [30] | Beccari L, Yakushiji-Kaminatsui N, Woltering JM, Necsulea A, Lonfat N, Rodríguez-Carballo E, Mascrez B, Yamamoto S, Kuroiwa A, Duboule D. A role for Hox13 proteins in the regulatory switch between TADs at the HoxD locus. Genes Dev, 2016, 30(10):1172-1186. |
| [31] | Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell, 2011, 147(5):1132-1145. |
| [32] | Lonfat N, Montavon T, Darbellay F, Gitto S, Duboule D. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science, 2014, 346(6212):1004-1006. |
| [33] |
Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: Transcriptional enhancers in development and evolution. Cell, 2016, 167(5):1170-1187.
doi: 10.1016/j.cell.2016.09.018 |
| [34] |
Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet, 2019, 20(8):437-455.
doi: 10.1038/s41576-019-0128-0 pmid: 31086298 |
| [35] |
Kim S, Shendure J. Mechanisms of interplay between transcription factors and the 3D genome. Mol Cell, 2019, 76(2):306-319.
doi: 10.1016/j.molcel.2019.08.010 |
| [36] | Noordermeer D, Leleu M, Splinter E, Rougemont J, De Laat W, Duboule D. The dynamic architecture of Hox gene clusters. Science, 2011, 334(6053):222-225. |
| [37] |
Li JH, Shou J, Guo Y, Tang YX, Wu YH, Jia ZL, Zhai YN, Chen ZF, Xu Q, Wu Q. Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9. J Mol Cell Biol, 2015, 7(4):284-298.
doi: 10.1093/jmcb/mjv016 |
| [38] |
Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res, 2013, 23(4):465-472.
doi: 10.1038/cr.2013.45 |
| [39] |
Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science, 2014, 346(6213):1258096.
doi: 10.1126/science.1258096 |
| [40] | Liu PF, Wu Q. Probing 3D genome by CRISPR/Cas9. Hereditas(Beijing), 2020, 42(1):18-31. |
| 刘沛峰, 吴强. CRISPR/Cas9基因编辑在三维基因组研究中的应用. 遗传, 2020, 42(1):18-31. | |
| [41] | Li JH, Shou J, Wu Q. DNA fragment editing of genomes by CRISPR/Cas9. Hereditas(Beijing), 2015, 37(10):992-1002. |
| 李金环, 寿佳, 吴强. CRISPR/Cas9系统在基因组DNA片段编辑中的应用. 遗传, 2015, 37(10):992-1002. | |
| [42] |
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with tophat and cufflinks. Nat Protoc, 2012, 7(3):562-578.
doi: 10.1038/nprot.2012.016 pmid: 22383036 |
| [43] | Guo XQ, Chen FZ, Gao F, Li L, Liu K, You LJ, Hua C, Yang F, Liu WL, Peng CH, Wang LN, Yang XX, Zhou FY, Tong JW, Cai J, Li ZY, Wan B, Zhang L, Yang T, Zhang MW, Yang LL, Yang YW, Zeng WJ, Wang B, Wei XF, Xu X. CNSA: A data repository for archiving omics data. Database (Oxford), 2020; 2020: baaa055. |
| [44] | Chen FZ, You LJ, Yang F, Wang LN, Guo XQ, Gao F, Hua C, Tan C, Fang L, Shan RQ, Zeng WJ, Wang B, Wang R, Xu X, Wei XF. CNGBdb: China National Genebank Database. Hereditas(Beijing), 2020, 42(08):799-809. |
| 陈凤珍, 游丽金, 杨帆, 王丽娜, 郭学芹, 高飞, 华聪, 谈聪, 方林, 单日强, 曾文君, 王博, 王韧, 徐讯, 魏晓锋. CNGBdb: 国家基因库生命大数据平台. 遗传, 2020, 42(8):799-809. | |
| [45] |
Pearson JC, Lemons D, McGinnis W. ModulatingHox gene functions during animal body patterning. Nat Rev Genet, 2005, 6(12):893-904.
pmid: 16341070 |
| [46] |
Lonfat N, Duboule D. Structure, function and evolution of topologically associating domains (TADs) atHox loci. FEBS Lett, 2015, 589(20):2869-2876.
doi: 10.1016/j.febslet.2015.04.024 |
| [47] | Holland PW, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl, 1994, 125-133. |
| [48] |
Shou J, Li J, Liu Y, Wu Q. Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell, 2018, 71(4):498-509 e4.
doi: S1097-2765(18)30466-0 pmid: 30033371 |
| [49] |
Fu YF, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol, 2013, 31(9):822-826.
doi: 10.1038/nbt.2623 |
| [50] |
Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci USA, 2011, 108(33):13570-13575.
doi: 10.1073/pnas.1109873108 |
| [51] |
Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature, 2010, 466(7305):490-493.
doi: 10.1038/nature09158 |
| [52] |
Barolo S. Shadow enhancers: Frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays, 2012, 34(2):135-141.
doi: 10.1002/bies.201100121 pmid: 22083793 |
| [53] |
Buecker C, Wysocka J. Enhancers as information integration hubs in development: Lessons from genomics. Trends Genet, 2012, 28(6):276-284.
doi: 10.1016/j.tig.2012.02.008 |
| [54] |
Jolma A, Yin YM, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J. DNA- dependent formation of transcription factor pairs alters their binding specificity. Nature, 2015, 527(7578):384-388.
doi: 10.1038/nature15518 |
| [55] |
Wang N, Jia ZL, Wu Q. RFX5 regulates gene expression of the Pcdhα cluster.Hereditas(Beijing), 2020, 42(8):760-774.
doi: 10.16288/j.yczz.20-184 pmid: 32952112 |
|
王娜, 甲芝莲, 吴强. RFX5调控原钙粘蛋白α基因簇的表达. 遗传, 2020, 42(8):760-774.
doi: 10.16288/j.yczz.20-184 pmid: 32952112 |
|
| [56] | Malik S, Roeder RG. The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet, 2010, 11(11):761-772. |
| [57] |
Bolt CC, Duboule D. The regulatory landscapes of developmental genes. Development, 2020, 147(3): dev171736.
doi: 10.1242/dev.171736 |
| [58] |
Neijts R, Deschamps J. At the base of colinearHox gene expression: Cis-features and trans-factors orchestrating the initial phase of Hox cluster activation. Dev Biol, 2017, 428(2):293-299.
doi: S0012-1606(16)30864-8 pmid: 28728680 |
| [59] |
Xu DF, Ma RS, Zhang JH, Liu ZJ, Wu B, Peng JH, Zhai YN, Gong QG, Shi YY, Wu JH, Wu Q, Zhang ZY, Ruan K. Dynamic nature of CTCF tandem 11 zinc fingers in multivalent recognition of DNA as revealed by NMR spectroscopy. J Phys Chem Lett, 2018, 9(14):4020-4028.
doi: 10.1021/acs.jpclett.8b01440 |
| [1] | Xiuli Chen, Haiyan Huang, Qiang Wu. Targeted deletion of 5′HS2 enhancer of β-globin locus control region in K562 cells [J]. Hereditas(Beijing), 2022, 44(9): 783-797. |
| [2] | Siyuan Xu, Jia Shou, Qiang Wu. Additional evidence of HS5-1 enhancer eRNA PEARL for protocadherin alpha gene regulation [J]. Hereditas(Beijing), 2022, 44(8): 695-764. |
| [3] | Sihan Qi, Qilin Wang, Junyou Zhang, Qian Liu, Chunyan Li. The regulatory mechanisms by enhancers during cancer initiation and progression [J]. Hereditas(Beijing), 2022, 44(4): 275-288. |
| [4] | Xingqi Wan, Wanzhen Wei, Shengliang Guo, Yixiao Cui, Xueying Jing, Lujie Huang, Jie Ma. Functional analysis of the long-range regulatory element of BMP2 gene [J]. Hereditas(Beijing), 2022, 44(12): 1141-1147. |
| [5] | Cong Zhou, Qiangwei Zhou, Sheng Cheng, Guoliang Li. Research progress of CTCF in mediating 3D genome formation and regulating gene expression [J]. Hereditas(Beijing), 2021, 43(9): 816-821. |
| [6] | Xianglong He, Jinhuan Li, Qiang Wu. Combinatorial CRISPR inversions of CTCF sites in HOXD cluster reveal complex insulator function [J]. Hereditas(Beijing), 2021, 43(8): 758-774. |
| [7] | Qian Liu, Chunyan Li. The identification of enhancers and its application in cancer studies [J]. Hereditas(Beijing), 2020, 42(9): 817-831. |
| [8] | Zhongyong Qin, Xiao Shi, Pingping Cao, Ying Chu, Wei Guan, Nan Yang, He Cheng, Yujie Sun. The NOXA promoter could function as an active enhancer to regulate the expression of BCL2 in the apoptosis response [J]. Hereditas(Beijing), 2020, 42(11): 1110-1121. |
| [9] | Zhiqiang Wu, Zeyun Mi. Research progress of super enhancer in cancer [J]. Hereditas(Beijing), 2019, 41(1): 41-51. |
| [10] | Xiao Cheng,Qiong Yang,Zhendong Tan,Ya Tan,Hongzhou Pu,Xue Zhao,Shunhua Zhang,Li Zhu. The current research status of enhancer RNAs [J]. Hereditas(Beijing), 2017, 39(9): 784-797. |
| [11] | Juntao Li,Wei Zhao,Dandan Li,Jing Feng,Gui Ba,Tianzeng Song,Hongping Zhang. miR-101a targeting EZH2 promotes the differentiation of goat skeletal muscle satellite cells [J]. Hereditas(Beijing), 2017, 39(9): 828-836. |
| [12] | Changbin Sun, Xi Zhang. Advance in the research on super-enhancer [J]. Hereditas(Beijing), 2016, 38(12): 1056-1068. |
| [13] | FENG Jun LI Guang WANG Yi-Quan. Research progress of conserved non-coding elements in metazoan [J]. HEREDITAS, 2013, 35(1): 35-44. |
| [14] | GUO Xin-Jun. Properties comparing and evolutionary analysis of MEF2 of Homo sapiens based on bioinformatic methods [J]. HEREDITAS, 2011, 33(9): 975-981. |
| [15] | GAO Yun-Zhen, BO Yu-Chun. Progress of transcription factor CCAAT enhancer binding protein β [J]. HEREDITAS, 2011, 33(3): 198-206. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||