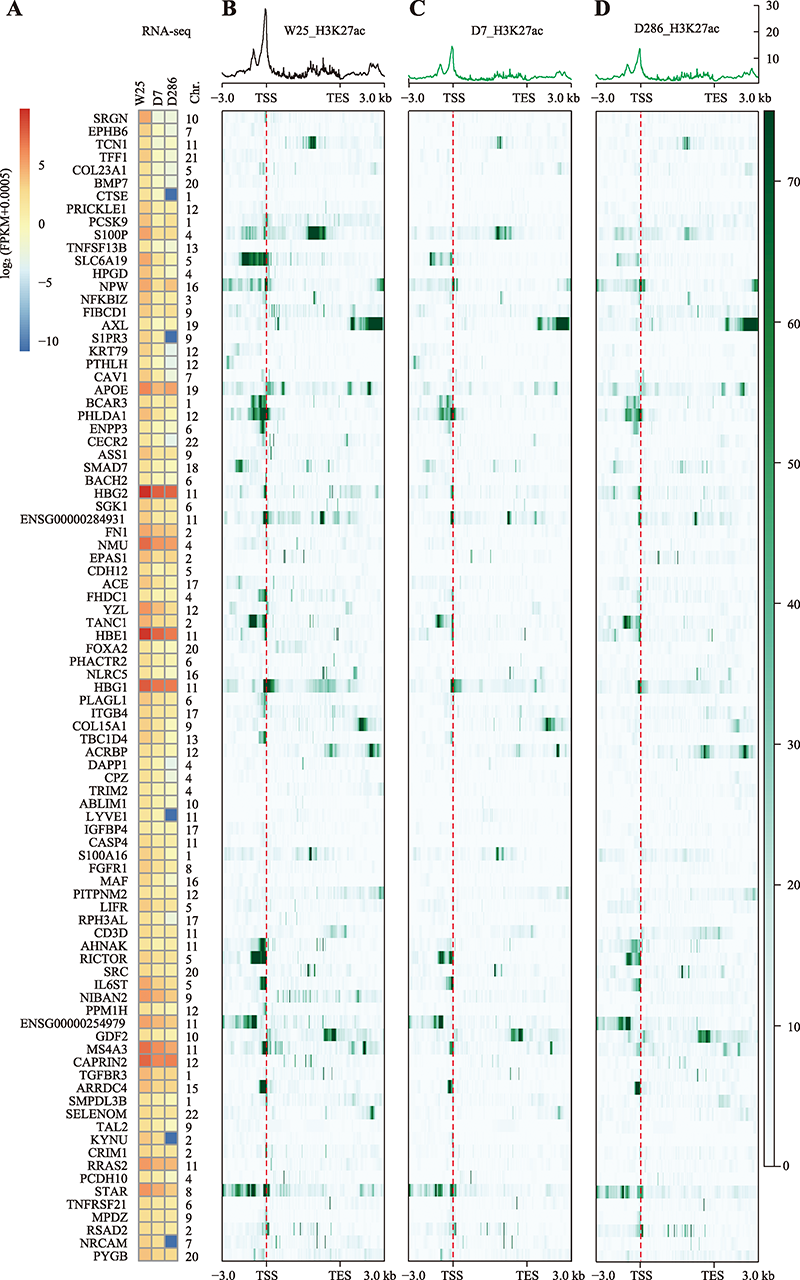
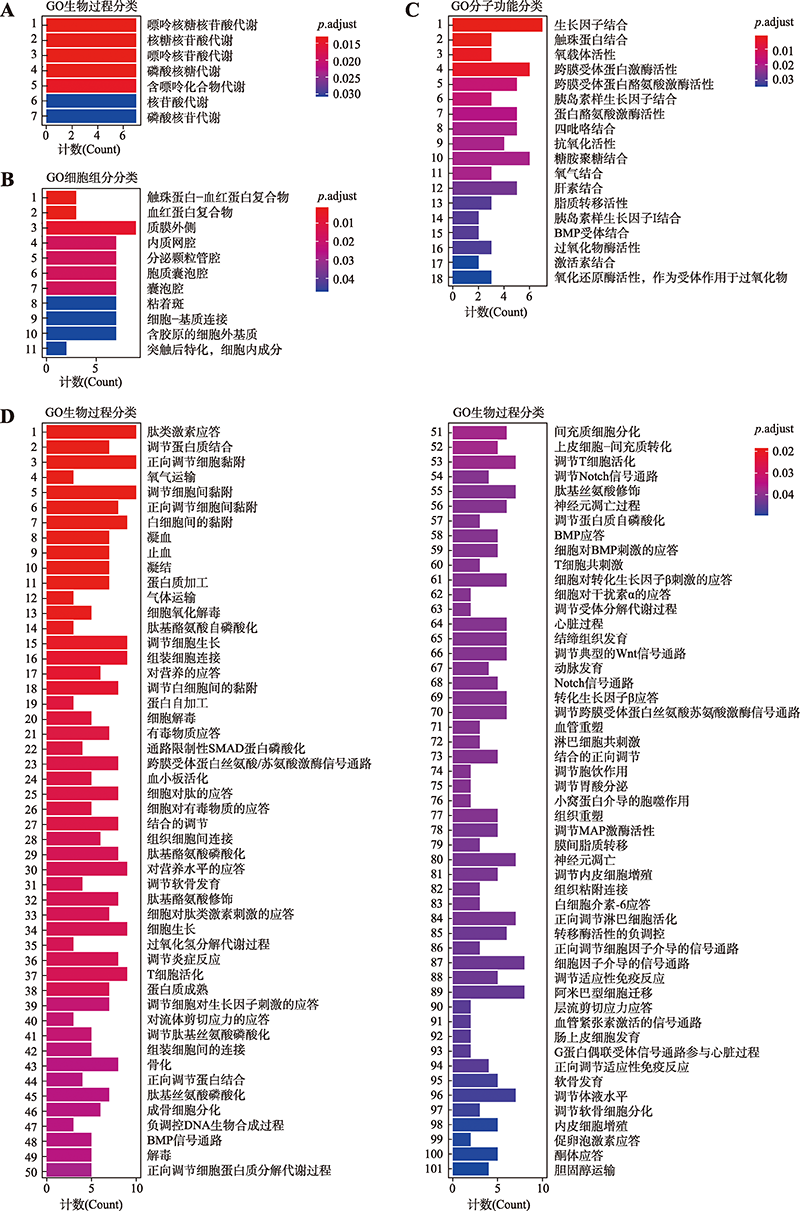
Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (9): 783-797.doi: 10.16288/j.yczz.22-201
• Research Article • Previous Articles Next Articles
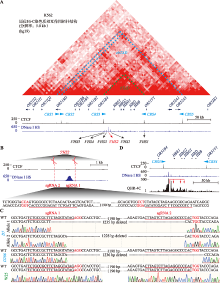
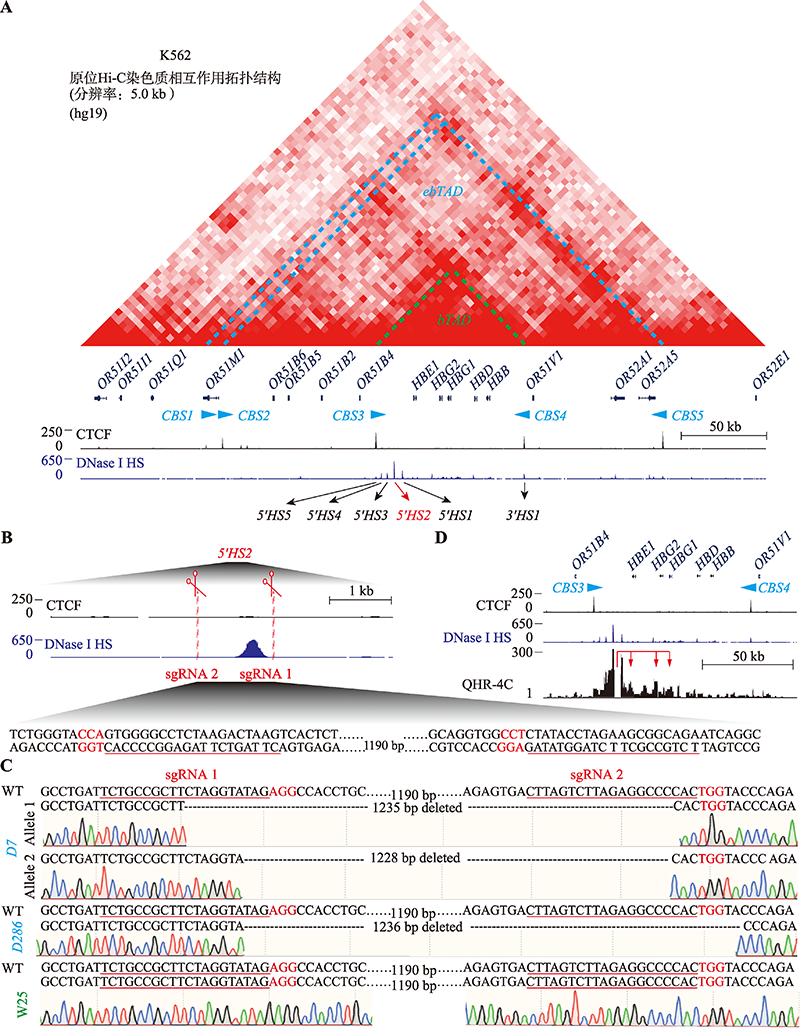
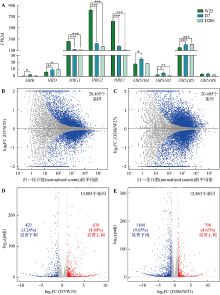
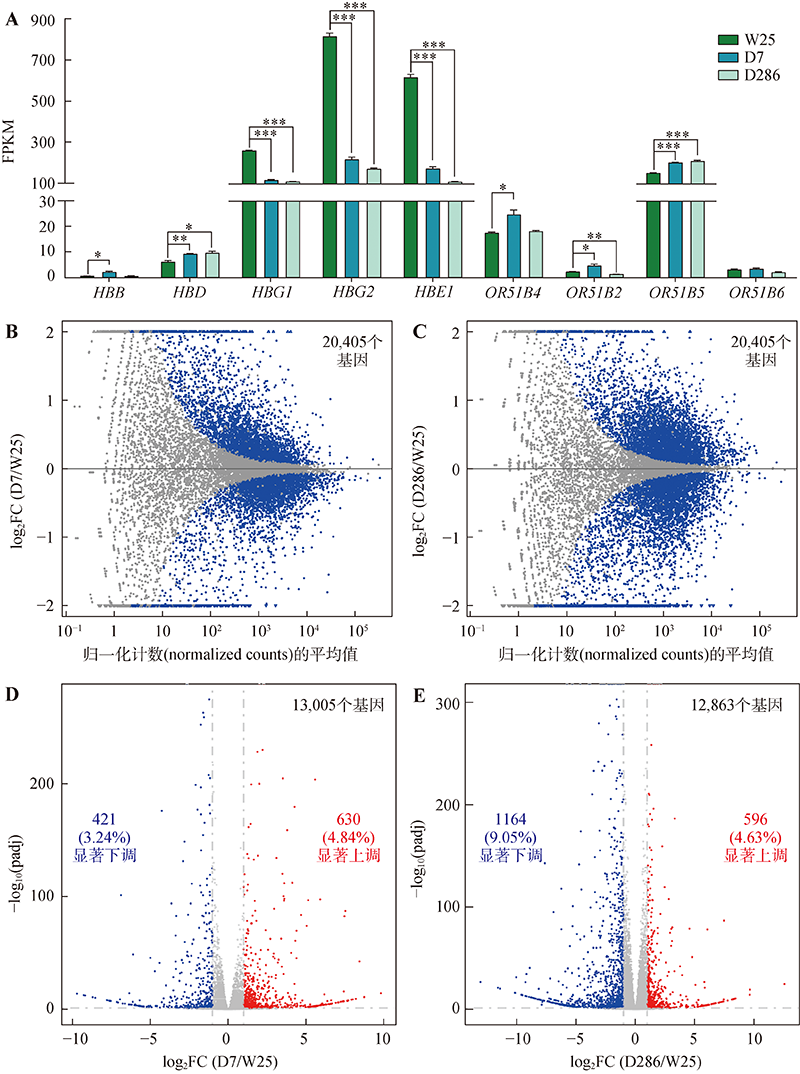
Targeted deletion of 5′HS2 enhancer of β-globin locus control region in K562 cells
Xiuli Chen( ), Haiyan Huang(
), Haiyan Huang( ), Qiang Wu(
), Qiang Wu( )
)
- Center for Comparative Biomedicine, Key laboratory of Systems Biomedicine (Ministry of Education), Institute of Systems Biomedicine, Shanghai Jiao Tong University, Shanghai 200240, China
-
Received:2022-06-16Revised:2022-07-20Online:2022-09-20Published:2022-08-01 -
Contact:Huang Haiyan,Wu Qiang E-mail:chenxiuli@sjtu.edu.cn;hy_huang@sjtu.edu.cn;qwu123@gmail.com -
Supported by:Supported by the National Natural Science Foundation of China(81872944);Supported by the National Natural Science Foundation of China(31630039);Science and Technology Commission of Shanghai Municipality Program Nos(19JC1412500);Science and Technology Commission of Shanghai Municipality Program Nos(21DZ2210200)
Cite this article
Xiuli Chen, Haiyan Huang, Qiang Wu. Targeted deletion of 5′HS2 enhancer of β-globin locus control region in K562 cells[J]. Hereditas(Beijing), 2022, 44(9): 783-797.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Primer sequences"
| 引物类型 | 引物名称 | 引物序列(5′→3′) |
|---|---|---|
| sgRNA | hHB_EHS2-sg1F | caccTCTGCCGCTTCTAGGTATAG |
| hHB_EHS2-sg1R | aaacCTATACCTAGAAGCGGCAGA | |
| hHB_EHS2-sg2F | caccCTTAGTCTTAGAGGCCCCAC | |
| hHB_EHS2-sg2R | aaacGTGGGGCCTCTAAGACTAAG | |
| PCR | universal-gRNA_R | GCACCGACTCGGTGCCACTT |
| hHB_EHS2- genotyping-F1 | GTAGTCCTTCACAGTTACCCACACA | |
| hHB_EHS2-genotyping-R1 | CATTAGTGACCTCCCATAGTCCAAG | |
| hHB_EHS2-genotyping-F2 | CCTGAGCTCCAAGCAGTCCAC | |
| hHB_EHS2-genotyping-R2 | CATTTGGCCCCTCCTAATCTCTC | |
| biotin修饰引物 | biotin-hs2[ | 5′biotin-CAGTTACCCACACAGGTGAACCC |
| P5引物 | qhr-hs2-F1[ | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGGAATGTTTCTTTCCTCTCAGGATC |
| P7引物 | wly-P2 | CAAGCAGAAGACGGCATACGAGATTTCATGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT |
| [1] | Hill RJ, Konigsberg W, Guidotti G, Craig LC . The structure of human hemoglobin. I. The separation of the alpha and beta chains and their amino acid composition. J Biol Chem, 1962,237:1549-1554. |
| [2] | Rahbar S, Beale D, Isaacs WA, Lehmann H . Abnormal haemoglobins in Iran. Observation of a new variant— haemoglobin J Iran (alpha-2-beta-2 77 His→Asp). Br Med J, 1967,1(5541):674-677. |
| [3] | Anson ML, Mirsky AE . On hemochromogen. J Gen Physiol, 1928,12(2):273-288. |
| [4] | Origa R . β-Thalassemia. Genet Med, 2017,19(6):609-619. |
| [5] | Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J . Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell, 1981,27(3 Pt 2):543-553. |
| [6] | Ingram VM . Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature, 1957,180(4581):326-328. |
| [7] | Grosveld F, Antoniou M, Berry M, De Boer E, Dillon N, Ellis J, Fraser P, Hanscombe O, Hurst J, Imam A, Lindenbaum M, Philipsen S, Pruzina S, Strouboulis J, Raguz-Bolognesi S, Talbot D . The regulation of human globin gene switching. Philos Trans R Soc Lond B Biol Sci, 1993,339(1288):183-191. |
| [8] | Guo Y, Xu Q, Canzio D, Shou J, Li JH, Gorkin DU, Jung I, Wu HY, Zhai YN, Tang YX, Lu YC, Wu YH, Jia ZL, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q . CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015,162(4):900-910. |
| [9] | Himadewi P, Wang XQD, Feng F, Gore H, Liu YS, Yu L, Kurita R, Nakamura Y, Pfeifer GP, Liu J, Zhang XT . 3′HS1 CTCF binding site in human β-globin locus regulates fetal hemoglobin expression. eLife, 2021,10:e70557. |
| [10] | Wai AWK, Gillemans N, Raguz-Bolognesi S, Pruzina S, Zafarana G, Meijer D, Philipsen S, Grosveld F . HS5 of the human beta-globin locus control region: a developmental stage-specific border in erythroid cells. EMBO J, 2003,22(17):4489-4500. |
| [11] | Pasceri P, Pannell D, Wu X, Ellis J . Full activity from human beta-globin locus control region transgenes requires 5′HS1, distal beta-globin promoter, and 3′ beta-globin sequences. Blood, 1998,92(2):653-663. |
| [12] | Fedosyuk H, Peterson KR . Deletion of the human beta-globin LCR 5′HS4 or 5′HS1 differentially affects beta-like globin gene expression in beta-YAC transgenic mice. Blood Cells Mol Dis, 2007,39(1):44-55. |
| [13] | Peterson KR, Clegg CH, Navas PA, Norton EJ, Kimbrough TG, Stamatoyannopoulos G . Effect of deletion of 5'HS3 or 5′HS2 of the human beta-globin locus control region on the developmental regulation of globin gene expression in beta-globin locus yeast artificial chromosome transgenic mice. Proc Natl Acad Sci USA, 1996,93(13):6605-6609. |
| [14] | Navas PA, Peterson KR, Li Q, Skarpidi E, Rohde A, Shaw SE, Clegg CH, Asano H, Stamatoyannopoulos G . Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol Cell Biol, 1998,18(7):4188-4196. |
| [15] | Kim S, Kim YW, Shim SH, Kim CG, Kim A . Chromatin structure of the LCR in the human β-globin locus transcribing the adult δ- and β-globin genes. Int J Biochem Cell Biol, 2012,44(3):505-513. |
| [16] | Navas PA, Peterson KR, Li Q, McArthur M, Stamatoyannopoulos G. The 5′HS4 core element of the human beta-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J Mol Biol, 2001,312(1):17-26. |
| [17] | Zhou YX, Xu SY, Zhang M, Wu Q . Systematic functional characterization of antisense eRNA of protocadherin α composite enhancer. Genes Dev, 2021,35(19-20):1383-1394. |
| [18] | Lee JH, Wang RY, Xiong F, Krakowiak J, Liao ZA, Nguyen PT, Moroz-Omori EV, Shao JF, Zhu XY, Bolt MJ, Wu HY, Singh PK, Bi MJ, Shi CJ, Jamal N, Li GJ, Mistry R, Jung SY, Tsai KL, Ferreon JC, Stossi F, Caflisch A, Liu ZJ, Mancini MA, Li WB. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell, 2021, 81(16): 3368-3385.e9. |
| [19] | Gurumurthy A, Yu DT, Stees JR, Chamales P, Gavrilova E, Wassel P, Li L, Stribling D, Chen JY, Brackett M, Ishov AM, Xie MY, Bungert J . Super-enhancer mediated regulation of adult β-globin gene expression: the role of eRNA and Integrator. Nucleic Acids Res, 2021,49(3):1383-1396. |
| [20] | Cho Y, Song SH, Lee JJ, Choi N, Kim CG, Dean A, Kim A . The role of transcriptional activator GATA-1 at human beta-globin HS2. Nucleic Acids Res, 2008,36(14):4521-4528. |
| [21] | Kang YJ, Kim YW, Kang J, Yun WJ, Kim A . Erythroid specific activator GATA-1-dependent interactions between CTCF sites around the β-globin locus. Biochim Biophys Acta Gene Regul Mech, 2017,1860(4):416-426. |
| [22] | Kim J, Kang J, Kim YW, Kim A . The human β-globin enhancer LCR HS2 plays a role in forming a TAD by activating chromatin structure at neighboring CTCF sites. FASEB J, 2021,35(6):e21669. |
| [23] | Jia ZL, Li JW, Ge X, Wu YH, Guo Y, Wu Q . Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol, 2020,21(1):75. |
| [24] | Martin M . Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J, 2011,17(1):10-12. |
| [25] | Kim D, Langmead B, Salzberg SL . HISAT: a fast spliced aligner with low memory requirements. Nat Methods, 2015,12(4):357-360. |
| [26] | Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol, 2010,28(5):511-515. |
| [27] | Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL . StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol, 2015,33(3):290-295. |
| [28] | Love MI, Huber W, Anders S . Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014,15(12):550. |
| [29] | Wu TZ, Hu EQ, Xu SB, Chen MJ, Guo PF, Dai ZH, Feng TZ, Zhou L, Tang WL, Zhan L, Fu XC, Liu SS, Bo XC, Yu GC. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb), 2021,2(3):100141. |
| [30] | Chen L, Zhang YH, Wang SP, Zhang YH, Huang T, Cai YD . Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One, 2017,12(9):e0184129. |
| [31] | Langmead B, Salzberg SL . Fast gapped-read alignment with Bowtie 2. Nat Methods, 2012,9(4):357-359. |
| [32] | Grytten I, Rand KD, Nederbragt AJ, Storvik GO, Glad IK, Sandve GK . Graph Peak Caller: calling ChIP-seq peaks on graph-based reference genomes. PLoS Comput Biol, 2019,15(2):e1006731. |
| [33] | Guo Y, Monahan K, Wu HY, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q . CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci USA, 2012,109(51):21081-21086. |
| [34] | Ziebarth JD, Bhattacharya A, Cui Y . CTCFBSDB 2.0: a database for CTCF-binding sites and genome organization. Nucleic Acids Res, 2013,41(Database issue):D188-D194. |
| [35] | Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL . A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 2014,159(7):1665-1680. |
| [36] | Wu Q, Shou J . Toward precise CRISPR DNA fragment editing and predictable 3D genome engineering. J Mol Cell Biol, 2021,12(11):828-856. |
| [37] | Hawkins RD, Hon GC, Yang CH, Antosiewicz-Bourget JE, Lee LK, Ngo QM, Klugman S, Ching KA, Edsall LE, Ye Z, Kuan S, Yu PZ, Liu H, Zhang XM, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Ren B . Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res, 2011,21(10):1393-1409. |
| [38] | Wang ZB, Zang CZ, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui KR, Roh TY, Peng WQ, Zhang MQ, Zhao KJ . Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet, 2008,40(7):897-903. |
| [39] | Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G . Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet, 2000,25(1):25-29. |
| [40] | Klein E, Ben-Bassat H, Neumann H, Ralph P, Zeuthen J, Polliack A, Vánky F . Properties of the K562 cell line, derived from a patient with chronic myeloid leukemia. Int J Cancer, 1976,18(4):421-431. |
| [41] | Andersson LC, Nilsson K, Gahmberg CG . K562--a human erythroleukemic cell line. Int J Cancer, 1979,23(2):143-147. |
| [42] | Cheng ML, Ho HY, Tseng HC, Lee CH, Shih LY, Chiu DTY . Antioxidant deficit and enhanced susceptibility to oxidative damage in individuals with different forms of alpha-thalassaemia. Br J Haematol, 2005,128(1):119-127. |
| [43] | Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo YQ, Wei CL, Ruan YJ, Bieker JJ, Fraser P . Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet, 2010,42(1):53-61. |
| [1] | Siyuan Xu, Jia Shou, Qiang Wu. Additional evidence of HS5-1 enhancer eRNA PEARL for protocadherin alpha gene regulation [J]. Hereditas(Beijing), 2022, 44(8): 695-764. |
| [2] | Sihan Qi, Qilin Wang, Junyou Zhang, Qian Liu, Chunyan Li. The regulatory mechanisms by enhancers during cancer initiation and progression [J]. Hereditas(Beijing), 2022, 44(4): 275-288. |
| [3] | Xingqi Wan, Wanzhen Wei, Shengliang Guo, Yixiao Cui, Xueying Jing, Lujie Huang, Jie Ma. Functional analysis of the long-range regulatory element of BMP2 gene [J]. Hereditas(Beijing), 2022, 44(12): 1141-1147. |
| [4] | Ling Wang, Jinhuan Li, Haiyan Huang, Qiang Wu. Serial deletions of tandem reverse CTCF sites reveal balanced HOXD regulatory landscape of enhancers [J]. Hereditas(Beijing), 2021, 43(8): 775-791. |
| [5] | Qian Liu, Chunyan Li. The identification of enhancers and its application in cancer studies [J]. Hereditas(Beijing), 2020, 42(9): 817-831. |
| [6] | Zhongyong Qin, Xiao Shi, Pingping Cao, Ying Chu, Wei Guan, Nan Yang, He Cheng, Yujie Sun. The NOXA promoter could function as an active enhancer to regulate the expression of BCL2 in the apoptosis response [J]. Hereditas(Beijing), 2020, 42(11): 1110-1121. |
| [7] | Peifeng Liu, Qiang Wu. Probing 3D genome by CRISPR/Cas9 [J]. Hereditas(Beijing), 2020, 42(1): 18-31. |
| [8] | Ke Yang, Zheng Xue, Xiang Lv. Molecular mechanism of the 3D genome structure and function regulation during cell terminal differentiation [J]. Hereditas(Beijing), 2020, 42(1): 32-44. |
| [9] | Zhiqiang Wu, Zeyun Mi. Research progress of super enhancer in cancer [J]. Hereditas(Beijing), 2019, 41(1): 41-51. |
| [10] | Xiao Cheng,Qiong Yang,Zhendong Tan,Ya Tan,Hongzhou Pu,Xue Zhao,Shunhua Zhang,Li Zhu. The current research status of enhancer RNAs [J]. Hereditas(Beijing), 2017, 39(9): 784-797. |
| [11] | Juntao Li,Wei Zhao,Dandan Li,Jing Feng,Gui Ba,Tianzeng Song,Hongping Zhang. miR-101a targeting EZH2 promotes the differentiation of goat skeletal muscle satellite cells [J]. Hereditas(Beijing), 2017, 39(9): 828-836. |
| [12] | Changbin Sun, Xi Zhang. Advance in the research on super-enhancer [J]. Hereditas(Beijing), 2016, 38(12): 1056-1068. |
| [13] | FENG Jun LI Guang WANG Yi-Quan. Research progress of conserved non-coding elements in metazoan [J]. HEREDITAS, 2013, 35(1): 35-44. |
| [14] | GUO Xin-Jun. Properties comparing and evolutionary analysis of MEF2 of Homo sapiens based on bioinformatic methods [J]. HEREDITAS, 2011, 33(9): 975-981. |
| [15] | GAO Yun-Zhen, BO Yu-Chun. Progress of transcription factor CCAAT enhancer binding protein β [J]. HEREDITAS, 2011, 33(3): 198-206. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||