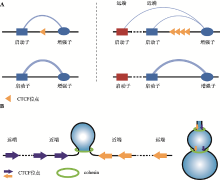
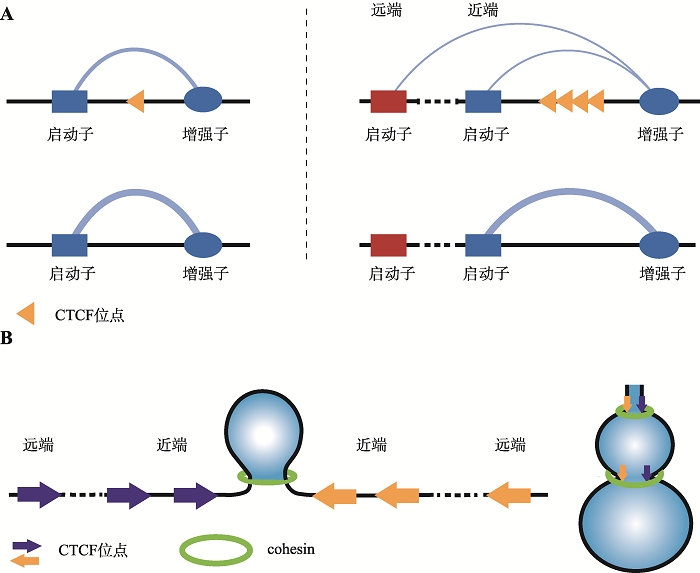
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (9): 816-821.doi: 10.16288/j.yczz.21-326
• Frontier Focus • Previous Articles Next Articles
Research progress of CTCF in mediating 3D genome formation and regulating gene expression
Cong Zhou( ), Qiangwei Zhou, Sheng Cheng, Guoliang Li(
), Qiangwei Zhou, Sheng Cheng, Guoliang Li( )
)
- Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China
-
Received:2021-09-08Revised:2021-09-13Online:2021-09-20Published:2021-09-15 -
Contact:Li Guoliang E-mail:zhoucong@mail.hzau.edu.cn;guoliang.li@mail.hzau.edu.cn -
Supported by:Supported by the National Natural Science Foundation of China Nos(31970590);Supported by the National Natural Science Foundation of China Nos(31771402)
Cite this article
Cong Zhou, Qiangwei Zhou, Sheng Cheng, Guoliang Li. Research progress of CTCF in mediating 3D genome formation and regulating gene expression[J]. Hereditas(Beijing), 2021, 43(9): 816-821.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Schmitt AD, Hu M, Ren B. Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol, 2016, 17(12):743-755.
doi: 10.1038/nrm.2016.104 |
| [2] |
Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet, 2019, 20(8):437-455.
doi: 10.1038/s41576-019-0128-0 pmid: 31086298 |
| [3] |
Zheng H, Xie W. The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol, 2019, 20(9):535-550.
doi: 10.1038/s41580-019-0132-4 |
| [4] |
Niu LJ, Shen W, Shi ZY, Tan YJ, He N, Wan J, Sun JL, Zhang YD, Huang YZ, Wang WJ, Fang C, Li JS, Zheng PP, Cheung E, Chen YL, Li L, Hou CH. Three-dimensional folding dynamics of the Xenopus tropicalis genome. Nat Genet, 2021, 53(7):1075-1087.
doi: 10.1038/s41588-021-00878-z |
| [5] |
Sanborn AL, Rao SSP, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, Geeting KP, Gnirke A, Melnikov A, McKenna D, Stamenova EK, Lander ES, Aiden EL. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci USA, 2015, 112(47):E6456-6465.
doi: 10.1073/pnas.1518552112 |
| [6] |
Guo Y, Xu Q, Canzio D, Shou J, Li JH, Gorkin DU, Jung I, Wu HY, Zhai YN, Tang YX, Lu YC, Wu YH, Jia ZL, Li W, Zhang MQ, Ren B, Krainer AR, Maniatis T, Wu Q. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell, 2015, 162(4):900-910.
doi: 10.1016/j.cell.2015.07.038 pmid: 26276636 |
| [7] |
Kim Y, Shi ZB, Zhang HS, Finkelstein IJ, Yu HT. Human cohesin compacts DNA by loop extrusion. Science, 2019, 366(6471):1345-1349.
doi: 10.1126/science.aaz4475 |
| [8] |
Handoko L, Xu H, Li GL, Ngan CY, Chew E, Schnapp M, Lee CWH, Ye CP, Ping JLH, Mulawadi F, Wong E, Sheng JP, Zhang YB, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan YJ, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet, 2011, 43(7):630-638.
doi: 10.1038/ng.857 pmid: 21685913 |
| [9] |
Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell, 2009, 137(7):1194-1211.
doi: 10.1016/j.cell.2009.06.001 |
| [10] |
Huang H, Zhu Q, Jussila A, Han YY, Bintu B, Kern C, Conte M, Zhang YX, Bianco S, Chiariello AM, Yu M, Hu R, Tastemel M, Juric I, Hu M, Nicodemi M, Zhuang XW, Ren B. CTCF mediates dosage- and sequence-context- dependent transcriptional insulation by forming local chromatin domains. Nat Genet, 2021, 53(7):1064-1074.
doi: 10.1038/s41588-021-00863-6 |
| [11] |
Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol, 1996, 16(6):2802-2813.
doi: 10.1128/MCB.16.6.2802 pmid: 8649389 |
| [12] |
Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell, 2015, 161(5):1012-1025.
doi: S0092-8674(15)00377-3 pmid: 25959774 |
| [13] |
Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature, 2011, 479(7371):74-79.
doi: 10.1038/nature10442 |
| [14] |
Vostrov AA, Quitschke WW. The zinc finger protein CTCF binds to the APBbeta domain of the amyloid beta-protein precursor promoter. Evidence for a role in transcriptional activation. J Biol Chem, 1997, 272(52):33353-33359.
pmid: 9407128 |
| [15] |
Zhang XF, Zhang Y, Ba ZQ, Kyritsis N, Casellas R, Alt FW. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature, 2019, 575(7782):385-389.
doi: 10.1038/s41586-019-1723-0 |
| [16] |
Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature, 2000, 405(6785):482-485.
doi: 10.1038/35013100 |
| [17] |
Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5'-flanking sequence of the chicken c-myc gene. Oncogene, 1990, 5(12):1743-1753.
pmid: 2284094 |
| [18] |
Jia ZL, Li JW, Ge X, Wu YH, Guo Y, Wu Q. Tandem CTCF sites function as insulators to balance spatial chromatin contacts and topological enhancer-promoter selection. Genome Biol, 2020, 21(1):75.
doi: 10.1186/s13059-020-01984-7 |
| [19] |
Tang ZH, Luo OJ, Li XW, Zheng MZ, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, Michalski P, Piecuch E, Wang P, Wang DJ, Tian SZ, Penrad-Mobayed M, Sachs LM, Ruan XA, Wei CL, Liu ET, Wilczynski GM, Plewczynski D, Li GL, Ruan YJ. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell, 2015, 163(7):1611-1627.
doi: 10.1016/j.cell.2015.11.024 |
| [20] |
Hyle J, Zhang Y, Wright S, Xu BS, Shao Y, Easton J, Tian LQ, Feng RP, Xu P, Li CL. Acute depletion of CTCF directly affects MYC regulation through loss of enhancer- promoter looping. Nucleic Acids Res, 2019, 47(13):6699-6713.
doi: 10.1093/nar/gkz462 |
| [21] |
Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell, 2017, 169(5): 930-944.e22.
doi: 10.1016/j.cell.2017.05.004 |
| [22] |
Xu BS, Wang H, Wright S, Hyle J, Zhang Y, Shao Y, Niu MM, Fan YP, Rosikiewicz W, Djekidel MN, Peng JM, Lu R, Li CL. Acute depletion of CTCF rewires genome-wide chromatin accessibility. Genome Biol, 2021, 22(1):244.
doi: 10.1186/s13059-021-02466-0 |
| [23] |
Kemp CJ, Moore JM, Moser R, Bernard B, Teater M, Smith LE, Rabaia NA, Gurley KE, Guinney J, Busch SE, Shaknovich R, Lobanenkov VV, Liggitt D, Shmulevich I, Melnick A, Filippova GN. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep, 2014, 7(4):1020-1029.
doi: 10.1016/j.celrep.2014.04.004 |
| [24] |
West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev, 2002, 16(3):271-288.
doi: 10.1101/gad.954702 |
| [25] | Wang L, Li JH, Huang HY, Wu Q. Serial deletions of tandem reverse CTCF sites reveal balanced HOXD regulatory landscape of enhancers. Hereditas(Beijing), 2021, 43(8):775-791. |
| 王玲, 李金环, 黄海燕, 吴强. 串联反向CTCF位点的系列删除揭示增强子调控HOXD基因簇表达的平衡. 遗传, 2021, 43(8):775-791. | |
| [26] | He XL, Li JH, Wu Q. Combinatorial CRISPR inversions of CTCF sites in HOXD cluster reveal complex insulator function. Hereditas(Beijing), 2021, 43(8):758-774. |
| 何象龙, 李金环, 吴强. HOXD基因簇内一系列CTCF位点反转揭示绝缘子功能. 遗传, 2021, 43(8):758-774. |
| [1] | Shunze Wang, Feng Jiang, Dongli Zhu, Tie-Lin Yang, Yan Guo. Application of Hi-C technology in three-dimensional genomics research and disease pathogenesis analysis [J]. Hereditas(Beijing), 2023, 45(4): 279-294. |
| [2] | Xianglong He, Jinhuan Li, Qiang Wu. Combinatorial CRISPR inversions of CTCF sites in HOXD cluster reveal complex insulator function [J]. Hereditas(Beijing), 2021, 43(8): 758-774. |
| [3] | Ling Wang, Jinhuan Li, Haiyan Huang, Qiang Wu. Serial deletions of tandem reverse CTCF sites reveal balanced HOXD regulatory landscape of enhancers [J]. Hereditas(Beijing), 2021, 43(8): 775-791. |
| [4] | Ye Wei, Ke Li, Daru Lu, Huaxing Zhu. Optimization of CUT&Tag product recovery and library construction method [J]. Hereditas(Beijing), 2021, 43(4): 362-374. |
| [5] | Na Wang, Zhilian Jia, Qiang Wu. RFX5 regulates gene expression of the Pcdhα cluster [J]. Hereditas(Beijing), 2020, 42(8): 760-774. |
| [6] | Yu Zhang, Yuda Fang. Progresses on the structure and function of cohesin [J]. Hereditas(Beijing), 2020, 42(1): 57-72. |
| [7] | Xiaofei Zheng,Haiyan Huang,Qiang Wu. Chromatin architectural protein CTCF regulates gene expression of the UGT1 cluster [J]. Hereditas(Beijing), 2019, 41(6): 509-523. |
| [8] | Dejian Xie, Minglei Shi, Yan Zhang, Tianyi Wang, Wenlong Shen, Bingyu Ye, Ping Li, Chao He, Xiangyuan Zhang, Zhihu Zhao. Construction of CTCF degradation cell line by CRISPR/Cas9 mediated genome editing [J]. HEREDITAS(Beijing), 2016, 38(7): 651-657. |
| [9] | Yanan Zhai, Quan Xu, Ya Guo, Qiang Wu. Characterization of a cluster of CTCF-binding sites in a protocadherin regulatory region [J]. HEREDITAS(Beijing), 2016, 38(4): 323-336. |
| [10] | MENG Ya-Nan, MENG Li-Jun, SONG E-Juan, LIU Mei-Ling, ZHANG Xiu-Jun. Small RNA molecules and regulation of spermatogenesis [J]. HEREDITAS, 2011, 33(1): 9-16. |
| [11] | SUN Ye-Ying, DENG Xiao-Jian, LV Yan, DONG Chun-Ling, WANG Ping-Rong, HUANG Xiao-Qun. Progress in Regulation of Rice Wx Gene Expression [J]. HEREDITAS, 2005, 27(6): 1013-1019. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||