Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (1): 46-62.doi: 10.16288/j.yczz.23-224
• Research Article • Previous Articles Next Articles
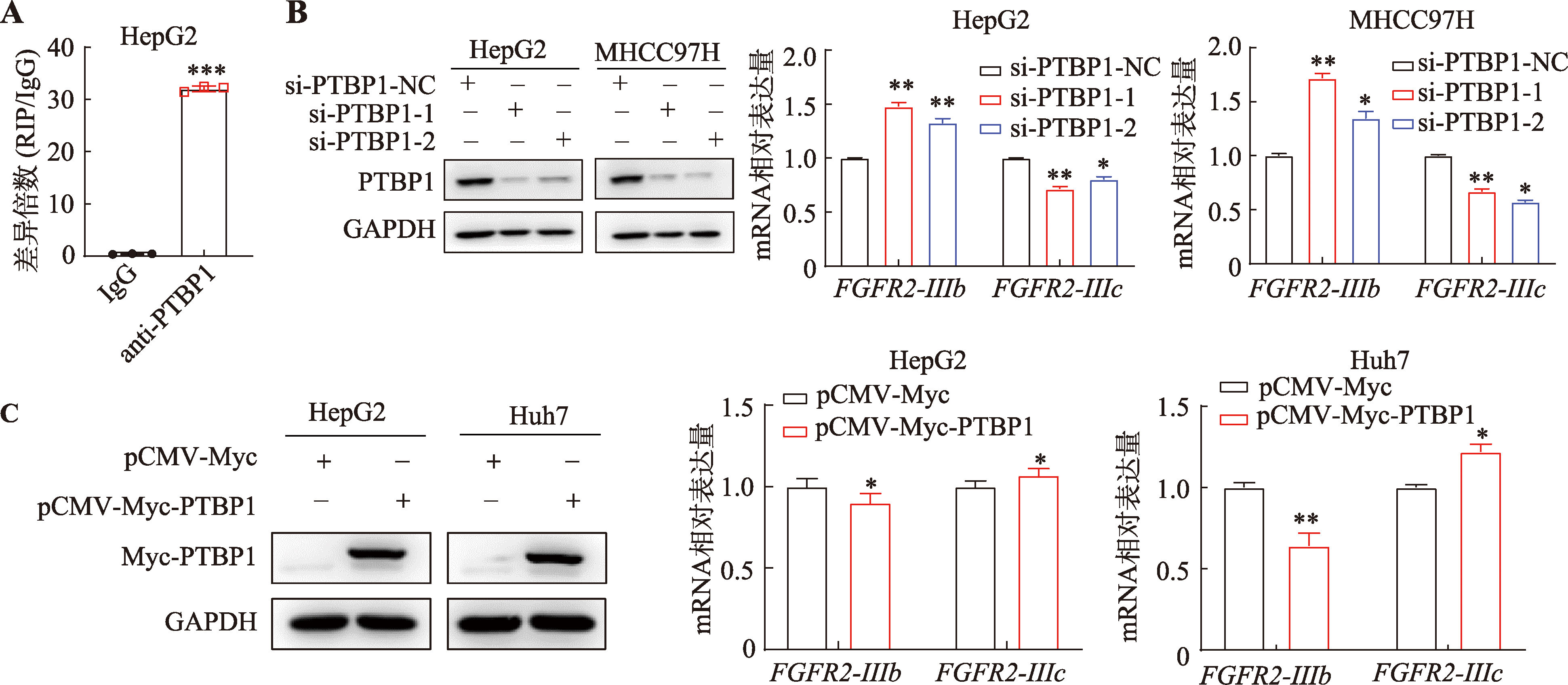
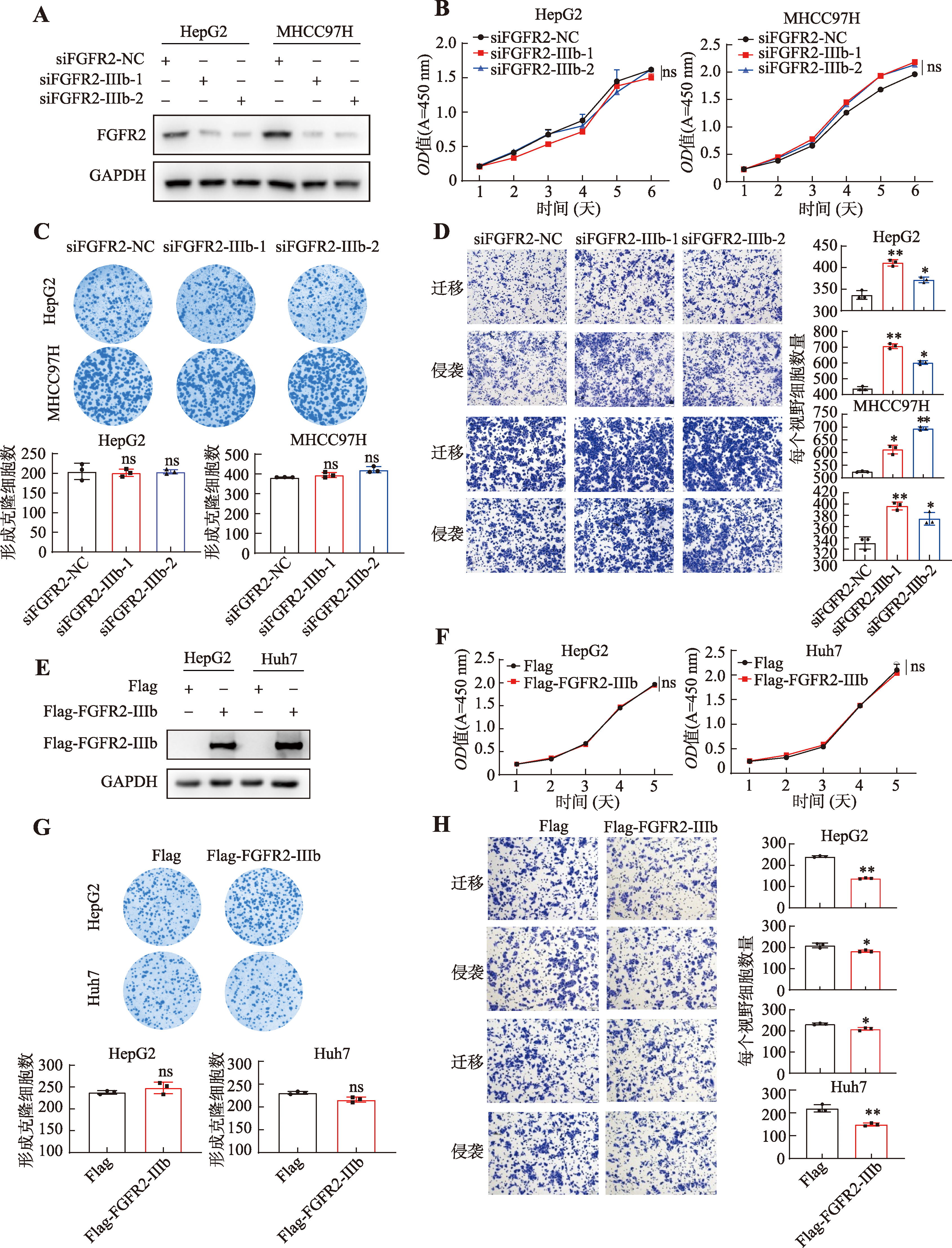
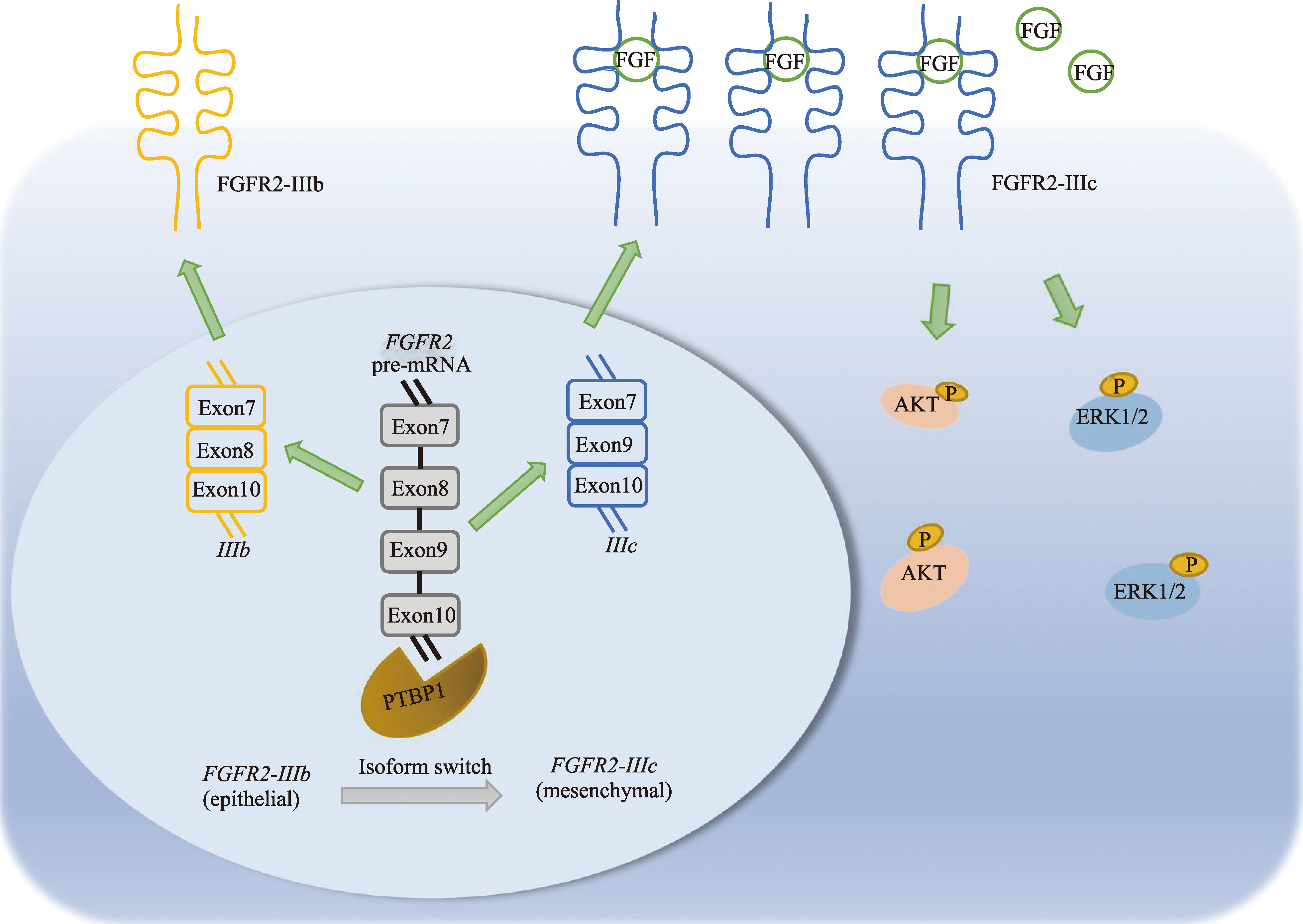
PTBP1 promotes the progression of hepatocellular carcinoma by enhancing the oncogenic splicing switch of FGFR2
Yuying Chen1,2( ), Qian Zhang3(
), Qian Zhang3( ), Menghui Gui4, Lan Feng2, Pengbo Cao2, Gangqiao Zhou1,2,4(
), Menghui Gui4, Lan Feng2, Pengbo Cao2, Gangqiao Zhou1,2,4( )
)
- 1. Hengyang Medical College, University of South China, Hengyang 421001, China
2. State Key Lab of Medical Proteomics, National Center for Protein Sciences at Beijing, Institute of Radiation Medicine, Academy of Military Medical Sciences, Academy of Military Sciences, Beijing 100850, China
3. National Facility for Translational Medicine, West China Hospital of Sichuan University, Chengdu 610041, China
4. School of Public Health, Nanjing Medical University, Nanjing 211166, China
-
Received:2023-08-23Revised:2023-11-08Online:2024-01-20Published:2023-12-05 -
Contact:Gangqiao Zhou E-mail:529897537@qq.com;15111174004@163.com;zhougq114@126.com -
Supported by:National Natural Science Foundation of China(82002573);National Natural Science Foundation of China(82172707);National Key Research and Development Program of China(2017YFA0504301)
Cite this article
Yuying Chen, Qian Zhang, Menghui Gui, Lan Feng, Pengbo Cao, Gangqiao Zhou. PTBP1 promotes the progression of hepatocellular carcinoma by enhancing the oncogenic splicing switch of FGFR2[J]. Hereditas(Beijing), 2024, 46(1): 46-62.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
The pathways and biological processes enriched by the interactors of PTBP1"
| 显著富集的信号通路及生物学 过程 | 富集的蛋白质数量 | 错误发现率 | 信号通路显著富集的蛋白质 |
|---|---|---|---|
| mRNA splicing-major pathway | 23 | 9.12E-20 | CPSF7、CSTF1、EIF4A3、FUS、HNRNPA1、HNRNPC、HNRNPD、HNRNPH2、HSPA8、NUDT21、PCBP1、PCBP2、POLR2A、POLR2B、PRPF19、PTBP1、PUF60、SF1、SF3B4、SNRNP40、SNW1、U2AF2、YBX1 |
| Processing of capped intron- containing pre-mRNA | 25 | 9.12E-20 | CPSF7、CSTF1、EIF4A3、FUS、HNRNPA1、HNRNPC、HNRNPD、HNRNPH2、HSPA8、NUDT21、NXF1、PCBP1、PCBP2、POLDIP3、POLR2A、POLR2B、PRPF19、PTBP1、PUF60、SF1、SF3B4、SNRNP40SNW1、U2AF2、YBX1 |
| rRNA processing in the nucleus and cytosol | 15 | 6.05E-10 | DDX21、EBNA1BP2、FBL、GNL3、NOP2、PES1、RPL10、RPL18、RPL28、RPL3、RPL36A、RPL4、RPL6、RPLP0、RRP1 |
| Major pathway of rRNA processing in the nucleolus and cytosol | 14 | 2.71E-09 | DDX21、EBNA1BP2、FBL、GNL3、PES1、RPL10、RPL18、RPL28、RPL3、RPL36A、RPL4、RPL6、RPLP0、RRP1 |
| FGFR2 alternative splicing | 7 | 1.02E-07 | HNRNPA1、POLR2A、POLR2B、PTBP1、RBFOX2、TIA1、TIAL1 |
| L13a-mediated translational silencing of ceruloplasmin expression | 10 | 2.93E-07 | EIF2S2、EIF2S3、RPL10、RPL18、RPL28、RPL3、RPL36A、RPL4、RPL6、RPLP0 |
| GTP hydrolysis and joining of the 60S ribosomal subunit | 10 | 2.93E-07 | EIF2S2、EIF2S3、RPL10、RPL18、RPL28、RPL3、RPL36A、RPL4、RPL6、RPLP0 |
| Influenza infection | 11 | 3.79E-07 | EIF2AK2、POLR2A、POLR2B、RPL10、RPL18、RPL28、RPL3、RPL36A、RPL4、RPL6、RPLP0 |
| Influenza viral RNA transcription and replication | 10 | 9.28E-07 | POLR2A、POLR2B、RPL10、RPL18、RPL28、RPL3、RPL36A、RPL4、RPL6、RPLP0 |
| [1] |
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2021, 71(3): 209-249.
doi: 10.3322/caac.v71.3 |
| [2] | Qiu HB, Cao SM, Xu RH. Cancer incidence, mortality, and burden in China: a time-trend analysis and comparison with the United States and United Kingdom based on the global epidemiological data released in 2020. Cancer Commun (Lond), 2021, 41(10): 1037-1048. |
| [3] |
Steyerberg EW, Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J, 2014, 35(29): 1925-1931.
doi: 10.1093/eurheartj/ehu207 pmid: 24898551 |
| [4] |
Guan Z, Cheng W, Huang D, Wei A. High MYBL2 expression and transcription regulatory activity is associated with poor overall survival in patients with hepatocellular carcinoma. Curr Res Transl Med, 2018, 66(1): 27-32.
doi: S2452-3186(17)30049-1 pmid: 29274707 |
| [5] | Tang R, Wu JC, Zheng LM, Li ZR, Zhou KL, Zhang ZS, Xu DF, Chen C. Long noncoding RNA RUSC1-AS-N indicates poor prognosis and increases cell viability in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci, 2018, 22(2): 388-396. |
| [6] |
Cheung HC, Hai T, Zhu W, Baggerly KA, Tsavachidis S, Krahe R, Cote GJ. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain, 2009, 132(Pt 8): 2277-2288.
doi: 10.1093/brain/awp153 pmid: 19506066 |
| [7] |
Takahashi H, Nishimura J, Kagawa Y, Kano Y, Takahashi Y, Wu X, Hiraki M, Hamabe A, Konno M, Haraguchi N, Takemasa I, Mizushima T, Ishii M, Mimori K, Ishii H, Doki Y, Mori M, Yamamoto H. Significance of polypyrimidine tract-binding protein 1 expression in colorectal cancer. Mol Cancer Ther, 2015, 14(7): 1705-1716.
doi: 10.1158/1535-7163.MCT-14-0142 pmid: 25904505 |
| [8] |
Ferrarese R, Harsh GR 4th, Yadav AK, Bug E, Maticzka D, Reichardt W, Dombrowski SM, Miller TE, Masilamani AP, Dai FP, Kim H, Hadler M, Scholtens DM, Yu ILY, Beck J, Srinivasasainagendra V, Costa F, Baxan N, Pfeifer D, von Elverfeldt D, Backofen R, Weyerbrock A, Duarte CW, He XL, Prinz M, Chandler JP, Vogel H, Chakravarti A, Rich JN, Carro MS, Bredel M,. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J Clin Invest, 2014, 124(7): 2861-2876.
doi: 10.1172/JCI68836 pmid: 24865424 |
| [9] |
Kang H, Heo S, Shin JJ, Ji E, Tak H, Ahn S, Lee KJ, Lee EK, Kim W. A miR-194/PTBP1/CCND3 axis regulates tumor growth in human hepatocellular carcinoma. J Pathol, 2019, 249(3): 395-408.
doi: 10.1002/path.v249.3 |
| [10] |
Wu DG, He XT, Wang WJ, Hu XT, Wang KF, Wang MH. Long noncoding RNA SNHG12 induces proliferation, migration, epithelial-mesenchymal transition, and stemness of esophageal squamous cell carcinoma cells via post-transcriptional regulation of BMI1 and CTNNB1. Mol Oncol, 2020, 14(9): 2332-2351.
doi: 10.1002/1878-0261.12683 pmid: 32239639 |
| [11] |
Li T, Jin XZ, Dong JR, Deng HB. Long noncoding RNA ARSR is associated with a poor prognosis in patients with colorectal cancer. J Gene Med, 2020, 22(10): e3241.
doi: 10.1002/jgm.v22.10 |
| [12] |
Wei L, Wang XW, Lv LY, Liu JB, Xing HX, Song YM, Xie MY, Lei TS, Zhang NS, Yang M. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer, 2019, 18(1): 147.
doi: 10.1186/s12943-019-1086-z pmid: 31651347 |
| [13] |
Gao G, Dai C, Yu X, Yin XB, Zhou F. Long noncoding RNA LINC00324 exerts protumorigenic effects on liver cancer stem cells by upregulating fas ligand via PU box binding protein. FASEB J, 2020, 34(4): 5800-5817.
doi: 10.1096/fj.201902705RR pmid: 32128906 |
| [14] |
Khadirnaikar S, Chatterjee A, Kumar P, Shukla S. A greedy algorithm-based stem cell lncRNA signature identifies a novel subgroup of lung adenocarcinoma patients with poor pognosis. Front Oncol, 2020, 10: 1203.
doi: 10.3389/fonc.2020.01203 pmid: 32850350 |
| [15] |
Lu GM, Rong YX, Liang ZJ, Hunag DL, Ma YF, Luo ZZ, Wu FX, Liu XH, Liu Y, Mo S, Qi ZQ, Li HM. Multiomics global landscape of stemness-related gene clusters in adipose-derived mesenchymal stem cells. Stem Cell Res Ther, 2020, 11(1): 310.
doi: 10.1186/s13287-020-01823-3 |
| [16] |
Sun LK, Wang L, Chen TX, Shi Y, Yao BW, Liu ZK, Wang YF, Li Q, Liu RK, Niu YS, Tu KS, Liu QG. LncRNA RUNX1-IT1 which is downregulated by hypoxia-driven histone deacetylase 3 represses proliferation and cancer stem-like properties in hepatocellular carcinoma cells. Cell Death Dis, 2020, 11(2): 95.
doi: 10.1038/s41419-020-2274-x pmid: 32024815 |
| [17] |
Shen LH, Lei SJ, Zhang B, Li SG, Huang LY, Czachor A, Breitzig M, Gao YM, Huang MY, Mo XM, Zheng Q, Sun HX, Wang F. Skipping of exon 10 in Axl pre-mRNA regulated by PTBP1 mediates invasion and metastasis process of liver cancer cells. Theranostics, 2020, 10(13): 5719-5735.
doi: 10.7150/thno.42010 |
| [18] |
He ZT, Ni QH, Li XC, Zhao MY, Mo QG, Duo YS. PTBP1 promotes hepatocellular carcinoma progression by regulating the skipping of exon 9 in NUMB pre-mRNA. Heliyon, 2023, 9(6): e17387.
doi: 10.1016/j.heliyon.2023.e17387 |
| [19] |
Guo JC, Yang YJ, Zheng JF, Zhang JQ, Guo M, Yang X, Jiang XL, Xiang L, Li Y, Ping H, Zhuo L. Silencing of long noncoding RNA HOXA11-AS inhibits the Wnt signaling pathway via the upregulation of HOXA11 and thereby inhibits the proliferation, invasion, and self-renewal of hepatocellular carcinoma stem cells. Exp Mol Med, 2019, 51(11): 1-20.
doi: 10.1038/s12276-019-0343-y pmid: 31704909 |
| [20] |
Amann T, Bataille F, Spruss T, Dettmer K, Wild P, Liedtke C, Mühlbauer M, Kiefer P, Oefner PJ, Trautwein C, Bosserhoff AK, Hellerbrand C. Reduced expression of fibroblast growth factor receptor 2IIIb in hepatocellular carcinoma induces a more aggressive growth. Am J Pathol, 2010, 176(3): 1433-1442.
doi: 10.2353/ajpath.2010.090356 pmid: 20093481 |
| [21] |
Yang H, Jiang Z, Wang S, Zhao YB, Song XM, Xiao YF, Yang SM. Long non-coding small nucleolar RNA host genes in digestive cancers. Cancer Med, 2019, 8(18): 7693-7704.
doi: 10.1002/cam4.v8.18 |
| [22] |
Zhu QY, Yang HJ, Cheng P, Han Q. Bioinformatic analysis of the prognostic value of the lncRNAs encoding snoRNAs in hepatocellular carcinoma. Biofactors, 2019, 45(2): 244-252.
doi: 10.1002/biof.1478 pmid: 30537372 |
| [23] |
Adamczyk-Gruszka O, Horecka-Lewitowicz A, Gruszka J, Wawszczak-Kasza M, Strzelecka A, Lewitowicz P. FGFR-2 and epithelial-mesenchymal transition in endometrial cancer. J Clin Med, 2022, 11(18): 5416.
doi: 10.3390/jcm11185416 |
| [24] |
Maehara O, Suda G, Natsuizaka M, Shigesawa T, Kanbe G, Kimura M, Sugiyama M, Mizokami M, Nakai M, Sho T, Morikawa K, Ogawa K, Ohashi S, Kagawa S, Kinugasa H, Naganuma S, Okubo N, Ohnishi S, Takeda H, Sakamoto N. FGFR2 maintains cancer cell differentiation via AKT signaling in esophageal squamous cell carcinoma. Cancer Biol Ther, 2021, 22(5-6): 372-380.
doi: 10.1080/15384047.2021.1939638 |
| [25] |
Czaplinska D, Turczyk L, Grudowska A, Mieszkowska M, Lipinska AD, Skladanowski AC, Zaczek AJ, Romanska HM, Sadej R. Phosphorylation of RSK2 at Tyr 529 by FGFR2-p38 enhances human mammary epithelial cells migration. Biochim Biophys Acta, 2014, 1843(11): 2461-2470.
doi: 10.1016/j.bbamcr.2014.06.022 pmid: 25014166 |
| [26] |
Cai SY, Yang YM, Jia BH, Wu ZH, Zhang JG, Shen JX, Qiu GX. Transcriptome-wide sequencing reveals molecules and pathways involved in neurofibromatosis type I combined with spinal deformities. Spine (Phila Pa 1976), 2020, 45(9): E489-E498.
doi: 10.1097/BRS.0000000000003338 |
| [27] |
Chaffee BR, Hoang TV, Leonard MR, Bruney DG, Wagner BD, Dowd JR, Leone G, Ostrowski MC, Robinson ML. FGFR and PTEN signaling interact during lens development to regulate cell survival. Dev Biol, 2016, 410(2): 150-163.
doi: S0012-1606(15)30127-5 pmid: 26764128 |
| [28] |
Nagaraju GP., Dariya B, Kasa P, Peela S, El-Rayes BF. Epigenetics in hepatocellular carcinoma. Semin Cancer Biol, 2022, 86(Pt 3): 622-632.
doi: 10.1016/j.semcancer.2021.07.017 |
| [29] |
Yang Y, Ren PW, Liu XF, Sun XY, Zhang CF, Du XJ, Xing BC. PPP1R26 drives hepatocellular carcinoma progression by controlling glycolysis and epithelial-mesenchymal transition. J Exp Clin Cancer Res, 2022, 41(1): 101.
doi: 10.1186/s13046-022-02302-8 pmid: 35292107 |
| [30] | Zhao JX, Wang F, Xu ZR, Fan YM. The epigenetic effect on pre-mRNA alternative splicing. Hereditas (Beijing), 2014, 36(3): 248-255. |
| 赵金璇, 王芳, 徐峥嵘, 范怡梅. 表观遗传调控pre-mRNA的选择性剪接. 遗传, 2014, 36(3): 248-255. | |
| [31] |
Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM, Liu QS, Pugh TJ, Pedamallu CS, Hayes DN, Gray NS, Getz G, Wong KK, Haddad RI, Meyerson M, Hammerman PS. Inhibitor-sensitive FGFR2 and FGFR3 mutations in lung squamous cell carcinoma. Cancer Res, 2013, 73(16): 5195-5205.
doi: 10.1158/0008-5472.CAN-12-3950 pmid: 23786770 |
| [32] |
Taniguchi K, Sakai M, Sugito N, Kumazaki M, Shinohara H, Yamada N, Nakayama T, Ueda H, Nakagawa Y, Ito Y, Futamura M, Uno B, Otsuki Y, Yoshida K, Uchiyama K, Akao Y. PTBP1-associated microRNA-1 and -133b suppress the Warburg effect in colorectal tumors. Oncotarget, 2016, 7(14): 18940-18952.
doi: 10.18632/oncotarget.8005 pmid: 26980745 |
| [33] |
Xie RH, Chen X, Chen ZY, Huang M, Dong W, Gu P, Zhang JT, Zhou QH, Dong W, Han JL, Wang XS, Li H, Huang J, Lin TX. Polypyrimidine tract binding protein 1 promotes lymphatic metastasis and proliferation of bladder cancer via alternative splicing of MEIS2 and PKM. Cancer Lett, 2019, 449: 31-44.
doi: S0304-3835(19)30071-0 pmid: 30742945 |
| [34] |
Kuranaga Y, Sugito N, Shinohara H, Tsujino T, Taniguchi K, Komura K, Ito Y, Soga T, Akao Y. SRSF3, a splicer of the PKM gene, regulates cell growth and maintenance of cancer-specific energy metabolism in colon cancer cells. Int J Mol Sci, 2018, 19(10): 3012.
doi: 10.3390/ijms19103012 |
| [35] |
Miao H, Wu F, Li Y, Qin CY, Zhao YY, Xie MF, Dai HY, Yao H, Cai HY, Wang QH, Song X, Li L. MALAT1 modulates alternative splicing by cooperating with the splicing factors PTBP1 and PSF. Sci Adv, 2022, 8(51): eabq7289.
doi: 10.1126/sciadv.abq7289 |
| [36] |
Teles SP, Oliveira P, Ferreira M, Carvalho J, Ferreira P, Oliveira C. Integrated analysis of structural variation and RNA expression of FGFR2 and its splicing modulator ESRP1 highlight the ESRP1(amp)-FGFR2(norm)-FGFR2- IIIc(high) axis in diffuse gastric cancer. Cancers (Basel), 2019, 12(1): 70.
doi: 10.3390/cancers12010070 |
| [37] |
Warzecha CC, Shen SH, Xing Y, Carstens RP. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol, 2009, 6(5): 546-562.
doi: 10.4161/rna.6.5.9606 pmid: 19829082 |
| [38] |
Jones RB, Carstens RP, Luo Y, McKeehan WL. 5′- and 3′-terminal nucleotides in the FGFR2 ISAR splicing element core have overlapping roles in exon IIIb activation and exon IIIc repression. Nucleic Acids Res, 2001, 29(17): 3557-3565.
pmid: 11522825 |
| [39] |
Simarro M, Mauger D, Rhee K, Pujana MA, Kedersha NL, Yamasaki S, Cusick ME, Vidal M, Garcia-Blanco MA, Anderson P. Fas-activated serine/threonine phosphoprotein (FAST) is a regulator of alternative splicing. Proc Natl Acad Sci USA, 2007, 104(27): 11370-11375.
doi: 10.1073/pnas.0704964104 pmid: 17592127 |
| [40] |
Hovhannisyan RH, Warzecha CC, Carstens RP. Characterization of sequences and mechanisms through which ISE/ISS-3 regulates FGFR2 splicing. Nucleic Acids Res, 2006, 34(1): 373-385.
pmid: 16410617 |
| [41] |
Yoshino M, Ishiwata T, Watanabe M, Matsunobu T, Komine O, Ono Y, Yamamoto T, Fujii T, Matsumoto K, Tokunaga A, Naito Z. Expression and roles of keratinocyte growth factor and its receptor in esophageal cancer cells. Int J Oncol, 2007, 31(4): 721-728.
pmid: 17786302 |
| [42] |
Ishiwata T, Friess H, Büchler MW, Lopez ME, Korc M. Characterization of keratinocyte growth factor and receptor expression in human pancreatic cancer. Am J Pathol, 1998, 153(1): 213-222.
doi: 10.1016/S0002-9440(10)65562-9 pmid: 9665482 |
| [43] |
Kurban G, Ishiwata T, Kudo M, Yokoyama M, Sugisaki Y, Naito Z. Expression of keratinocyte growth factor receptor (KGFR/FGFR2 IIIb) in human uterine cervical cancer. Oncol Rep, 2004, 11(5): 987-991.
pmid: 15069536 |
| [44] |
Yamayoshi T, Nagayasu T, Matsumoto K, Abo T, Hishikawa Y, Koji T. Expression of keratinocyte growth factor/fibroblast growth factor-7 and its receptor in human lung cancer: correlation with tumour proliferative activity and patient prognosis. J Pathol, 2004, 204(1): 110-118.
pmid: 15307144 |
| [45] |
Cho K, Ishiwata T, Uchida E, Nakazawa N, Korc M, Naito Z, Tajiri T. Enhanced expression of keratinocyte growth factor and its receptor correlates with venous invasion in pancreatic cancer. Am J Pathol, 2007, 170(6): 1964-1974.
pmid: 17525264 |
| [46] |
Zhang Y, Wang H, Toratani S, Sato JD, Kan M, McKeehan WL, Okamoto T. Growth inhibition by keratinocyte growth factor receptor of human salivary adenocarcinoma cells through induction of differentiation and apoptosis. Proc Natl Acad Sci USA, 2001, 98(20): 11336-11340.
doi: 10.1073/pnas.191377098 pmid: 11562460 |
| [47] |
Matsubara A, Kan M, Feng S, McKeehan WL.Inhibition of growth of malignant rat prostate tumor cells by restoration of fibroblast growth factor receptor 2. Cancer Res, 1998, 58(7): 1509-1514.
pmid: 9537256 |
| [48] |
Ricol D, Cappellen D, El Marjou A, Gil-Diez-de-Medina S, Girault JM, Yoshida T, Ferry G, Tucker G, Poupon MF, Chopin D, Thiery JP, Radvanyi F. Tumour suppressive properties of fibroblast growth factor receptor 2-IIIb in human bladder cancer. Oncogene, 1999, 18(51): 7234-7243.
pmid: 10602477 |
| [49] |
Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol, 2006, 7(2): 131-142.
doi: 10.1038/nrm1835 |
| [50] |
Warzecha CC, Carstens RP. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin Cancer Biol, 2012, 22(5-6): 417-427.
doi: 10.1016/j.semcancer.2012.04.003 pmid: 22548723 |
| [51] |
Mittal V. Epithelial mesenchymal transition in tumor metastasis. Annu Rev Pathol, 2018, 13: 395-412.
doi: 10.1146/annurev-pathol-020117-043854 pmid: 29414248 |
| [52] |
Carstens RP, Eaton JV, Krigman HR, Walther PJ, Garcia-Blanco MA. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene, 1997, 15(25): 3059-3065.
pmid: 9444954 |
| [53] |
Zhao Q, Caballero OL, Davis ID, Jonasch E, Tamboli P, Yung WK, Weinstein JN, Kenna Shaw for TCGA research network, Strausberg RL, Yao J. Tumor-specific isoform switch of the fibroblast growth factor receptor 2 underlies the mesenchymal and malignant phenotypes of clear cell renal cell carcinomas. Clin Cancer Res, 2013, 19(9): 2460-2472.
doi: 10.1158/1078-0432.CCR-12-3708 pmid: 23444225 |
| [54] |
Huynh H, Ngo VC, Fargnoli J, Ayers M, Soo KC, Koong HN, Thng CH, Ong HS, Chung A, Chow P, Pollock P, Byron S, Tran E. Brivanib alaninate, a dual inhibitor of vascular endothelial growth factor receptor and fibroblast growth factor receptor tyrosine kinases, induces growth inhibition in mouse models of human hepatocellular carcinoma. Clin Cancer Res, 2008, 14(19): 6146-6153.
doi: 10.1158/1078-0432.CCR-08-0509 pmid: 18829493 |
| [1] | Min Cheng, Jing Zhang, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of the hypoxia-associated long non-coding RNA signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2022, 44(2): 153-167. |
| [2] | Hongbo Luo, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of a DNA methylation-driven transcriptional signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2020, 42(8): 775-787. |
| [3] | Xuqing Liu,Yubang Gao,Liangzhen Zhao,Yuchen Cai,Huiyuan Wang,Miao Miao,Lianfeng Gu,Hangxiao Zhang. Biogenesis, research methods, and functions of circular RNAs [J]. Hereditas(Beijing), 2019, 41(6): 469-485. |
| [4] | Qiying Leng, Jiahui Zheng, Haidong Xu, Patricia Adu-Asiamah, Ying Zhang, Bingwang Du, Li Zhang. Cloning and expression analysis of chicken circular transcript of insulin degrading enzyme gene [J]. Hereditas(Beijing), 2019, 41(12): 1129-1137. |
| [5] | Dong Wang, Yongjun Li, Nan Ding, Junyun Wang, Qiong Yang, Yaran Yang, Yanming Li, Xiangdong Fang, Hua Zhao. Molecular networks and mechanisms of epithelial-mesenchymal transition regulated by miRNAs in the malignant melanoma cell line [J]. HEREDITAS(Beijing), 2015, 37(7): 673-682. |
| [6] | Lingyun Sun, Xingyu Li, Zhiwei Sun. Progress of epigenetics and its therapeutic application in hepatocellular carcinoma [J]. HEREDITAS(Beijing), 2015, 37(6): 517-527. |
| [7] | Chengxiao Zhao, Ze Yang. Biological function and molecular mechanism of Twist2 [J]. HEREDITAS(Beijing), 2015, 37(1): 17-24. |
| [8] | Jiao Li, Yuqi Guo, Weiling Cui, Aihua Xu, Zengyuan Tian. Response of maize serine/arginine-rich protein gene family in seedlings to drought stress [J]. HEREDITAS(Beijing), 2014, 36(7): 697-706. |
| [9] | Lihua Yang, Xingkai Shen, Jingqiu Li, Jie Yang, Yanping Le, Zhaohui Gong. MicroRNAs affect tumor metastasis through regulating epithelial- mesenchymal transition [J]. HEREDITAS(Beijing), 2014, 36(7): 637-645. |
| [10] | Jinxuan Zhao, Fang Wang, Zhengrong Xu, Yimei Fan. The epigenetic effect on pre-mRNA alternative splicing [J]. HEREDITAS, 2014, 36(3): 248-255. |
| [11] | LUO Yan, WANG Ying. Alternative splicing of SmU2AFin Solanum melongena L. [J]. HEREDITAS, 2008, 30(11): 1499-1505. |
| [12] | WANG Wei, LAI Mao-De. Alternative Splicing of Insulin Receptor mRNA in Cancer and Type 2 Diabetes Mellitus: A Review [J]. HEREDITAS, 2006, 28(2): 226-230. |
| [13] | ZHOU Li-Wei, MA Fei, LI Qing-Wei. Progress in the Study on Alternative Splicing and Functions of Kininogen Genes [J]. HEREDITAS, 2006, 28(12): 1649-1649~1655. |
| [14] | XU Xian-Guo, WU Jun-Jie, HONG Xiao-Zhen, ZHU Fa-Ming, YAN Li-Xing. Identification of Nine Novel Alternative Splicing Isoforms of RHD mRNA [J]. HEREDITAS, 2006, 28(10): 1213-1218. |
| [15] | LIN Lu -Ping, MA Fei, WANG Yi-Quan. The Application of Bioinformatics in the Research of Alternative Splicing [J]. HEREDITAS, 2005, 27(6): 1001-1006. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||