Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (9): 716-726.doi: 10.16288/j.yczz.24-124
• Research Article • Previous Articles Next Articles
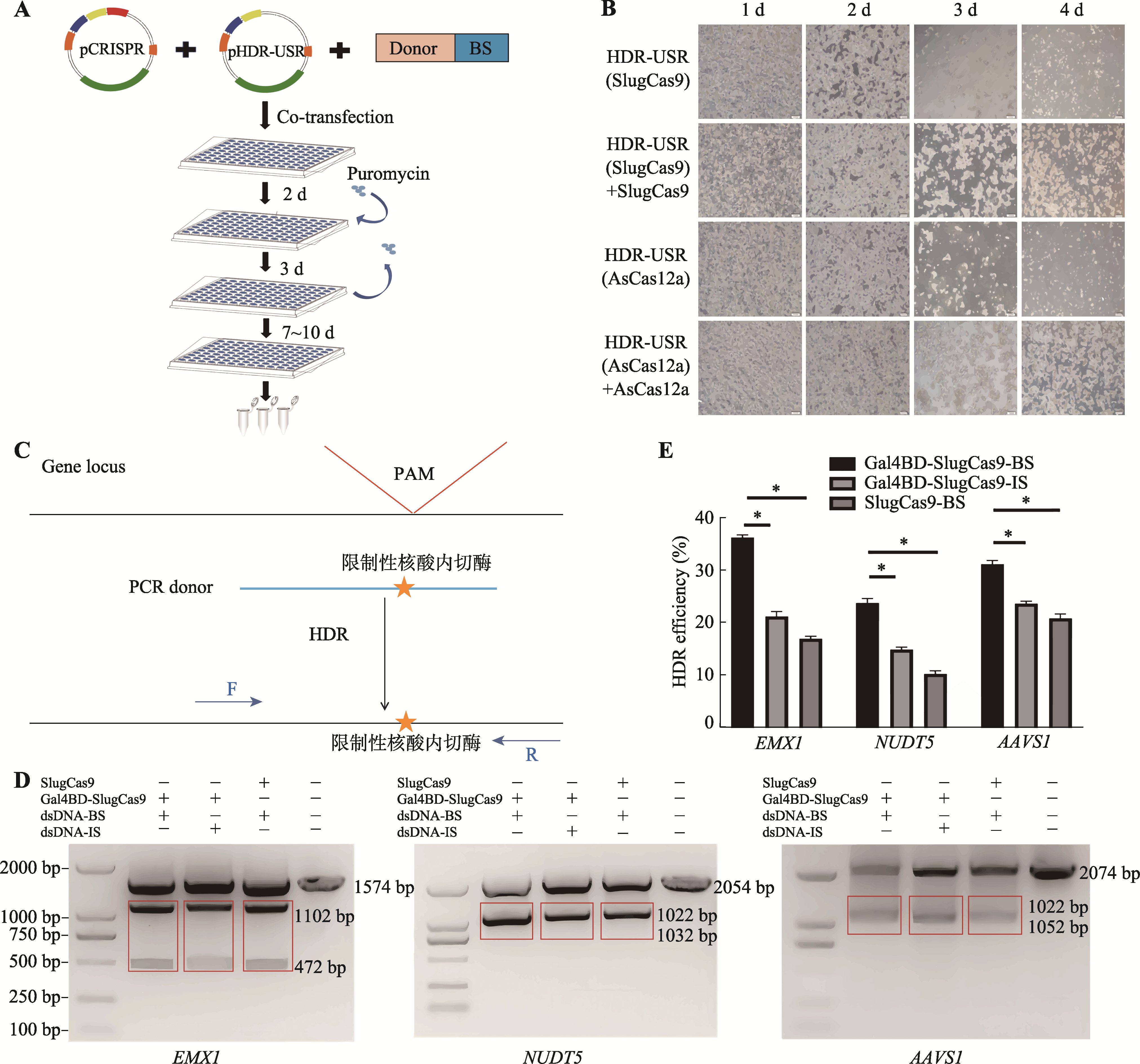
CRISPR/Gal4BD-Cas donor adapting systems based on miniaturized Cas proteins for improved gene editing
Sen Yang( ), Baoxia Ma, Hongrun Qian, Jieyu Cui, Xiaojun Zhang, Lida Li, Zehui Wei, Zhiying Zhang, Jiangang Wang(
), Baoxia Ma, Hongrun Qian, Jieyu Cui, Xiaojun Zhang, Lida Li, Zehui Wei, Zhiying Zhang, Jiangang Wang( ), Kun Xu(
), Kun Xu( )
)
- College of Animal Science and Technology, Northwest A&F University, Yangling 71200, China
-
Received:2024-06-28Revised:2024-08-24Online:2024-08-26Published:2024-08-26 -
Contact:Jiangang Wang, Kun Xu E-mail:yangsen@nwafu.edu.cn;wangjiangang@126.com;xukunas@nwafu.edu.cn -
Supported by:Biological Breeding-Major Projects(2023ZD04074);Biological Breeding-Major Projects(2023ZD04051)
Cite this article
Sen Yang, Baoxia Ma, Hongrun Qian, Jieyu Cui, Xiaojun Zhang, Lida Li, Zehui Wei, Zhiying Zhang, Jiangang Wang, Kun Xu. CRISPR/Gal4BD-Cas donor adapting systems based on miniaturized Cas proteins for improved gene editing[J]. Hereditas(Beijing), 2024, 46(9): 716-726.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Comparison of different miniaturized Cas proteins"
| 蛋白名称 | 蛋白来源 | 类型 | 蛋白长度 | PAM序列 | gRNA类型 | 参考文献 |
|---|---|---|---|---|---|---|
| SaCas9 | Staphylococcus aureus | II-A | 1053 aa | -NNGRRT | sgRNA | [ |
| SchCas9 | Staphylococcus chromogenes | II-A | 1054 aa | -NNGR | tracrRNA | [ |
| SlugCas9 | Staphylococcus lugdunensis | II-A | 1054 aa | -NNGG | sgRNA | [ |
| CjCas9 | Campylobacter jejuni | II-C | 984 aa | -NNNNRYAC | sgRNA | [ |
| NmeCas9 | N. meningitidis strain 8013 | II-C | 1081 aa | -NNNNGATT | sgRNA | [ |
| AsCas12a | Acidaminococcus | V-A | 1307 aa | TTTV- | crRNA | [ |
| LbCas12a | Lachnospiraceae bacterium | V-A | 1228 aa | TTTV- | crRNA | [ |
| Cas12c | − | V-C | 1209~1330 aa | 5′TG/5′TN | tracrRNA | [ |
| Cas12j(Casφ) | Biggiephage | V-J | 700~800 aa | TTN- | crRNA | [ |
| AsCas12f1 | Acidibacillus sulfuroxidans | V-F | 422 aa | NTTR- | sgRNA | [ |
| SpCas12f1 | Syntrophomonas palmitatica | V-F | 497 aa | TTC- | sgRNA | [ |
| CasMINI | Engineering from Un1Cas12f1 | V-F | 529 aa | TTTR- | sgRNA | [ |
| Cas12h | − | V-H | 870~924 aa | 5′RTR | crRNA | [ |
| Cas12i | − | V-I | 1033~1093 aa | 5′TTN | crRNA | [ |
| Cas12g | − | V-G | 720~830 aa | 无 | crRNA、tracrRNA | [ |
| [1] | Wu YX, Zeng J, Roscoe BP, Liu PP, Yao QM, Lazzarotto CR, Clement K, Cole MA, Luk K, Baricordi C, Shen AH, Ren CY, Esrick EB, Manis JP, Dorfman DM, Williams DA, Biffi A, Brugnara C, Biasco L, Brendel C, Pinello L, Tsai SQ, Wolfe SA, Bauer DE. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med, 2019, 25(5): 776-783. |
| [2] |
Chen YO, Bao Y, Ma HZ, Yi ZY, Zhou Z, Wei WS. Gene editing technology and its research progress in China. Hereditas(Beijing), 2018, 40(10): 900-915.
doi: 10.16288/j.yczz.18-195 pmid: 30369472 |
| 陈一欧, 宝颖, 马华峥, 伊宗裔, 周卓, 魏文胜. 基因编辑技术及其在中国的研究发展. 遗传, 2018, 40(10): 900-915. | |
| [3] |
Nambiar TS, Baudrier L, Billon P, Ciccia A. CRISPR- based genome editing through the lens of DNA repair. Mol Cell, 2022, 82(2): 348-388.
doi: 10.1016/j.molcel.2021.12.026 pmid: 35063100 |
| [4] | Scully R, Panday A, Elango R, Willis NA. DNA double- strand break repair-pathway choice in somatic mammalian cells. Nat Rev Mol Cell Biol, 2019, 20(11): 698-714. |
| [5] | Paix A, Folkmann A, Goldman DH, Kulaga H, Grzelak MJ, Rasoloson D, Paidemarry S, Green R, Reed RR, Seydoux G. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc Natl Acad Sci USA, 2017, 114(50): E10745-E10754. |
| [6] |
Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci USA, 2001, 98(15): 8403-8410.
doi: 10.1073/pnas.111009698 pmid: 11459982 |
| [7] |
Nambiar TS, Billon P, Diedenhofen G, Hayward SB, Taglialatela A, Cai KH, Huang JW, Leuzzi G, Cuella-Martin R, Palacios A, Gupta A, Egli D, Ciccia A. Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat Commun, 2019, 10(1): 3395.
doi: 10.1038/s41467-019-11105-z pmid: 31363085 |
| [8] |
Riesenberg S, Maricic T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun, 2018, 9(1): 2164.
doi: 10.1038/s41467-018-04609-7 pmid: 29867139 |
| [9] |
Wienert B, Nguyen DN, Guenther A, Feng SJ, Locke MN, Wyman SK, Shin J, Kazane KR, Gregory GL, Carter MAM, Wright F, Conklin BR, Marson A, Richardson CD, Corn JE. Timed inhibition of CDC7 increases CRISPR- Cas9 mediated templated repair. Nat Commun, 2020, 11(1): 2109.
doi: 10.1038/s41467-020-15845-1 pmid: 32355159 |
| [10] |
Ma M, Zhuang FF, Hu XB, Wang BL, Wen XZ, Ji JF, Jeff Xi J. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res, 2017, 27(4): 578-581.
doi: 10.1038/cr.2017.29 pmid: 28266543 |
| [11] |
Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat Commun, 2017, 8(1): 1711.
doi: 10.1038/s41467-017-01875-9 pmid: 29167458 |
| [12] |
Ali Z, Shami A, Sedeek K, Kamel R, Alhabsi A, Tehseen M, Hassan N, Butt H, Kababji A, Hamdan SM, Mahfouz MM. Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun Biol, 2020, 3(1): 44.
doi: 10.1038/s42003-020-0768-9 pmid: 31974493 |
| [13] |
Keegan L, Gill G, Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science, 1986, 231(4739): 699-704.
pmid: 3080805 |
| [14] | Giniger E, Ptashne M. Cooperative DNA binding of the yeast transcriptional activator GAL4. Proc Natl Acad Sci USA, 1988, 85(2): 382-386. |
| [15] | Zhang XJ, Xu K, Shen JC, Mu L, Qian HR, Cui JY, Ma BX, Chen ZL, Zhang ZY, Wei ZH. A CRISPR/Cas9-Gal4BD donor adapting system for enhancing homology-directed repair. Hereditas(Beijing), 2022, 44(8): 708-719. |
| 张潇筠, 徐坤, 沈俊岑, 穆璐, 钱泓润, 崔婕妤, 马宝霞, 陈知龙, 张智英, 魏泽辉. 一种新型提高HDR效率的CRISPR/Cas9-Gal4BD供体适配基因编辑系统. 遗传, 2022, 44(8): 708-719. | |
| [16] |
Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfò I, Maus MV, Pinello L, Aryee MJ, Joung JK. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol, 2019, 37(3): 276-282.
doi: 10.1038/s41587-018-0011-0 pmid: 30742127 |
| [17] |
Tóth E, Varga É, Kulcsár PI, Kocsis-Jutka V, Krausz SL, Nyeste A, Welker Z, Huszár K, Ligeti Z, Tálas A, Welker E. Improved LbCas12a variants with altered PAM specificities further broaden the genome targeting range of Cas12a nucleases. Nucleic Acids Res, 2020, 48(7): 3722-3733.
doi: 10.1093/nar/gkaa110 pmid: 32107556 |
| [18] | Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu XB, Makarova KS, Koonin EV, Sharp PA, Zhang F. In vivo genome editing using Staphylococcus aureus Cas9. Nature, 2015, 520(7546): 186-191. |
| [19] | Wang S, Mao HL, Hou LH, Hu ZY, Wang Y, Qi T, Tao C, Yang Y, Zhang CD, Li MM, Liu HH, Hu SJ, Chai RJ, Wang YM. Compact SchCas9 recognizes the simple NNGR PAM. Adv Sci (Weinh), 2022, 9(4): e2104789. |
| [20] |
Hu ZY, Zhang CD, Wang S, Gao SQ, Wei JJ, Li MM, Hou LH, Mao HL, Wei YY, Qi T, Liu HM, Liu D, Lan F, Lu DR, Wang HY, Li JX, Wang YM. Discovery and engineering of small SlugCas9 with broad targeting range and high specificity and activity. Nucleic Acids Res, 2021, 49(7): 4008-4019.
doi: 10.1093/nar/gkab148 pmid: 33721016 |
| [21] | Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim JS. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun, 2017, 8: 14500. |
| [22] | Hou ZG, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA, 2013, 110(39): 15644-15649. |
| [23] |
Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF, Doudna JA. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science, 2020, 369(6501): 333-337.
doi: 10.1126/science.abb1400 pmid: 32675376 |
| [24] |
Wu ZW, Zhang YF, Yu HP, Pan D, Wang YJ, Wang YN, Li F, Liu C, Nan H, Chen WZ, Ji QJ. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat Chem Biol, 2021, 17(11): 1132-1138.
doi: 10.1038/s41589-021-00868-6 pmid: 34475565 |
| [25] |
Bigelyte G, Young JK, Karvelis T, Budre K, Zedaveinyte R, Djukanovic V, Van Ginkel E, Paulraj S, Gasior S, Jones S, Feigenbutz L, Clair GS, Barone P, Bohn J, Acharya A, Zastrow-Hayes G, Henkel-Heinecke S, Silanskas A, Seidel R, Siksnys V. Miniature type V-F CRISPR-Cas nucleases enable targeted DNA modification in cells. Nat Commun, 2021, 12(1): 6191.
doi: 10.1038/s41467-021-26469-4 pmid: 34702830 |
| [26] | Kim DY, Lee JM, Moon SB, Chin HJ, Park S, Lim Y, Kim D, Koo T, Ko JH, Kim YS. Efficient CRISPR editing with a hypercompact Cas12f1 and engineered guide RNAs delivered by adeno-associated virus. Nat Biotechnol, 2022, 40(1): 94-102. |
| [27] |
Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston- Smith E, Sothiselvam S, Garrity AJ, Chong SR, Makarova KS, Koonin EV, Cheng DR, Scott DA. Functionally diverse type V CRISPR-Cas systems. Science, 2019, 363(6422): 88-91.
doi: 10.1126/science.aav7271 pmid: 30523077 |
| [28] |
Zhang LY, Zuris JA, Viswanathan R, Edelstein JN, Turk R, Thommandru B, Rube HT, Glenn SE, Collingwood MA, Bode NM, Beaudoin SF, Lele S, Scott SN, Wasko KM, Sexton S, Borges CM, Schubert MS, Kurgan GL, McNeill MS, Fernandez CA, Myer VE, Morgan RA, Behlke MA, Vakulskas CA. AsCas12a ultra nuclease facilitates the rapid generation of therapeutic cell medicines. Nat Commun, 2021, 12(1): 3908.
doi: 10.1038/s41467-021-24017-8 pmid: 34162850 |
| [29] |
Fagerlund RD, Staals RHJ, Fineran PC. The Cpf1 CRISPR-Cas protein expands genome-editing tools. Genome Biol, 2015, 16: 251.
doi: 10.1186/s13059-015-0824-9 pmid: 26578176 |
| [30] | Shao SM, Ren CH, Liu ZT, Bai YC, Chen ZL, Wei ZH, Wang X, Zhang ZY, Xu K. Enhancing CRISPR/ Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int J Biochem Cell Biol, 2017, 92: 43-52. |
| [31] | Xu K, Ren CH, Liu ZT, Zhang T, Zhang TT, Li D, Wang L, Yan Q, Guo LJ, Shen JC, Zhang ZY. Efficient genome engineering in eukaryotes using Cas9 from Streptococcus thermophilus. Cell Mol Life Sci, 2015, 72(2): 383-399. |
| [32] | Yan NN, Sun YS, Fang YY, Deng JR, Mu L, Xu K, Mymryk JS, Zhang ZY. A universal surrogate reporter for efficient enrichment of CRISPR/Cas9-mediated homology- directed repair in mammalian cells. Mol Ther Nucleic Acids, 2020, 19: 775-789. |
| [33] | Qian WD, Huang J, Wang XF, Wang T, Li YD. CRISPR-Cas12a combined with reverse transcription recombinase polymerase amplification for sensitive and specific detection of human norovirus genotype GII.4. Virology, 2021, 564: 26-32. |
| [34] | Zheng SY, Ma LL, Wang XL, Lu LX, Ma ST, Xu B, Ouyang W. RPA-Cas12aDS: A visual and fast molecular diagnostics platform based on RPA-CRISPR-Cas12a method for infectious bursal disease virus detection. J Virol Methods, 2022, 304: 114523. |
| [35] |
Nguyen DN, Roth TL, Li PJ, Chen PA, Apathy R, Mamedov MR, Vo LT, Tobin VR, Goodman D, Shifrut E, Bluestone JA, Puck JM, Szoka FC, Marson A. Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat Biotechnol, 2020, 38(1): 44-49.
doi: 10.1038/s41587-019-0325-6 pmid: 31819258 |
| [36] | Savic N, Ringnalda FC, Lindsay H, Berk C, Bargsten K, Li YZ, Neri D, Robinson MD, Ciaudo C, Hall J, Jinek M, Schwank G. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology- directed repair. eLife, 2018, 7: e33761. |
| [37] |
Ma SF, Wang XL, Hu YF, Lv J, Liu CF, Liao KT, Guo XH, Wang D, Lin Y, Rong ZL. Enhancing site-specific DNA integration by a Cas9 nuclease fused with a DNA donor-binding domain. Nucleic Acids Res, 2020, 48(18): 10590-10601.
doi: 10.1093/nar/gkaa779 pmid: 32986839 |
| [1] | Dongxia Pan, Hui Wang, Benhai Xiong, Xiangfang Tang. Progress on CRISPR-Cas gene editing technology in sheep production [J]. Hereditas(Beijing), 2024, 46(9): 690-700. |
| [2] | Ma Baoxia, Yang Sen, Lyu Ming, Wang Yuren, Chang Liye, Han Yifan, Wang Jiangang, Guo Yang, Xu Kun. Comparison and optimization of different CRISPR/Cas9 donor-adapting systems for gene editing [J]. Hereditas(Beijing), 2024, 46(6): 466-477. |
| [3] | Zhenlin Cao, Jinhong Li, Minhui Zhou, Manting Zhang, Ning Wang, Yifei Chen, Jiaxin Li, Qingsong Zhu, Wenjun Gong, Xuchen Yang, Xiaolong Fang, Jiaxian He, Meina Li. Functional study of the soybean stamen-preferentially expressed gene GmFLA22a in regulating male fertility [J]. Hereditas(Beijing), 2024, 46(4): 333-345. |
| [4] | Yanchun Bao, Lingli Dai, Zaixia Liu, Fengying Ma, Yu Wang, Yongbin Liu, Mingjuan Gu, Risu Na, Wenguang Zhang. Progress on CRISPR/Cas9 system in the genetic improvement of livestock and poultry [J]. Hereditas(Beijing), 2024, 46(3): 219-231. |
| [5] | Zhong Bian, Dongping Cao, Wenshu Zhuang, Shuwei Zhang, Qiaoquan Liu, Lin Zhang. Revelation of rice molecular design breeding: the blend of tradition and modernity [J]. Hereditas(Beijing), 2023, 45(9): 718-740. |
| [6] | Bingzheng Wang, Chao Zhang, Jiali Zhang, Jin Sun. Conditional editing of the Drosophila melanogaster genome using single transcripts expressing Cas9 and sgRNA [J]. Hereditas(Beijing), 2023, 45(7): 593-601. |
| [7] | Xiaojun Zhang, Kun Xu, Juncen Shen, Lu Mu, Hongrun Qian, Jieyu Cui, Baoxia Ma, Zhilong Chen, Zhiying Zhang, Zehui Wei. A CRISPR/Cas9-Gal4BD donor adapting system for enhancing homology-directed repair [J]. Hereditas(Beijing), 2022, 44(8): 708-719. |
| [8] | Yuting Han, Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni. The cutting edge of gene regulation approaches in model organism Drosophila [J]. Hereditas(Beijing), 2022, 44(1): 3-14. |
| [9] | Dingwei Peng, Ruiqiang Li, Wu Zeng, Min Wang, Xuan Shi, Jianhua Zeng, Xiaohong Liu, Yaoshen Chen, Zuyong He. Editing the cystine knot motif of MSTN enhances muscle development of Liang Guang Small Spotted pigs [J]. Hereditas(Beijing), 2021, 43(3): 261-270. |
| [10] | Yingnan Chen, Jing Lu. Application of CRISPR/Cas9 mediated gene editing in trees [J]. Hereditas(Beijing), 2020, 42(7): 657-668. |
| [11] | Guoling Li, Shanxin Yang, Zhenfang Wu, Xianwei Zhang. Recent developments in enhancing the efficiency of CRISPR/Cas9- mediated knock-in in animals [J]. Hereditas(Beijing), 2020, 42(7): 641-656. |
| [12] | Lianchao Tang, Feng Gu. Next-generation CRISPR-Cas for genome editing: focusing on the Cas protein and PAM [J]. Hereditas(Beijing), 2020, 42(3): 236-249. |
| [13] | Junxia Cao, Youliang Wang, Zhengxu Wang. Advances in precise regulation of CRISPR/Cas9 gene editing technology [J]. Hereditas(Beijing), 2020, 42(12): 1168-1177. |
| [14] | Qianqian Zhang,Xuan Shang,Wanying Lin,Xiangmin Xu. Effect of genetic modifiers on the clinical severity of β-thalassemia [J]. Hereditas(Beijing), 2019, 41(8): 669-676. |
| [15] | Xuran Niu,Shuming Yin,Xi Chen,Tingting Shao,Dali Li. Gene editing technology and its recent progress in disease therapy [J]. Hereditas(Beijing), 2019, 41(7): 582-598. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||