Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (8): 708-719.doi: 10.16288/j.yczz.22-118
• Research Article • Previous Articles Next Articles
A CRISPR/Cas9-Gal4BD donor adapting system for enhancing homology-directed repair
Xiaojun Zhang( ), Kun Xu(
), Kun Xu( ), Juncen Shen, Lu Mu, Hongrun Qian, Jieyu Cui, Baoxia Ma, Zhilong Chen, Zhiying Zhang, Zehui Wei(
), Juncen Shen, Lu Mu, Hongrun Qian, Jieyu Cui, Baoxia Ma, Zhilong Chen, Zhiying Zhang, Zehui Wei( )
)
- College of Animal Science and Technology, Northwest A&F University, Yangling 71200, China
-
Received:2022-04-05Revised:2022-05-11Online:2022-08-20Published:2022-05-23 -
Contact:Xu Kun,Wei Zehui E-mail:mshn15@163.com;weizehui7848@163.com;xukunas@nwafu.edu.cn -
Supported by:the National Natural Science Foundation of China(32172736);the Shaanxi Key R&D Program(2021NY-027)
Cite this article
Xiaojun Zhang, Kun Xu, Juncen Shen, Lu Mu, Hongrun Qian, Jieyu Cui, Baoxia Ma, Zhilong Chen, Zhiying Zhang, Zehui Wei. A CRISPR/Cas9-Gal4BD donor adapting system for enhancing homology-directed repair[J]. Hereditas(Beijing), 2022, 44(8): 708-719.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Comparison of different strategies to improve HDR efficiency"
| 分类 | 具体策略 | 检测所用细胞类型 | 不同类型供体提升效果 | 代表性 参考文献 | |
|---|---|---|---|---|---|
| dsDNA | ssDNA | ||||
| 抑制NHEJ 通路 | 抑制DNA连接酶4 | HEK293/K562/MEF/DC2.4/MelJuSo/HCT-116 | 2.4~19倍 | ~3倍 | [ |
| 抑制蛋白激酶催化亚基 | HEK293/iPSC | 2~5倍 | 1.6~3.3倍 | [ | |
| 抑制Ku蛋白 | HEK293/PFF | 2~3倍 | ~2.4倍 | [ | |
| 抑制53BP1 | HEK293/Hela/K562/U2OS/LCL B/iPSC | 1.4~3倍 | 1.3~3.3倍 | [ | |
| 增强HDR 通路 | 共表达CtIP转录因子 | HEK293/iPSC | 1.5~14.9倍 | — | [ |
| 共表达外切核酸酶1 | K562/A549/H1299 | 2~2.5倍 | — | [ | |
| 共表达Rad51、Rad52 | HEK293/PK15/iPSC | 2~6倍 | 1.4~2.4倍 | [ | |
| 优化供体 形式 | ssDNA供体优化 | HEK293/U2OS/T cells | — | ~3倍 | [ |
| 环状dsDNA供体优化 | HEK293/HeLa/MCF10A/Embryo | 1.1~18倍 | — | [ | |
| 线性dsDNA供体优化 | HEK293/iPSC | 2~10倍 | 1.4~10倍 | [ | |
| 供体与DBS共定位 | HEK293 | 3~6倍 | 1.6~18倍 | [ | |
| 控制打靶 时效 | 调节细胞周期 | HEK293/iPSC/T cells | 1.7~6倍 | — | [ |
| Cas9定时生效 | HEK293/Two-cell embryo | 1.87~10倍 | 1~5倍 | [ | |
| 其他机制 | 小分子化合物 | HEK293/PFF/iPSC | 2~3倍 | 2~10倍 | [ |
| 染色质状态 | PFF/hES/iPSC/Embryo | ~2倍 | 2-3倍 | [ | |
Table 2
Several CRISPR/Cas9-derived donor adapting systems"
| 系统简称 | 融合端 | Linker | 适配配体 | 配体大小 | 适配受体 | 受体大小 | 供体形式 | 参考文献 |
|---|---|---|---|---|---|---|---|---|
| Cas9-SNAP/ ssDNA | Cas9 C-terminus | Undeclared | SNAP-tag | 181 aa | O6-BG | ~241 (MW) | ssDNA | [ |
| Cas9-Avidin/ dsDNA | Cas9 | SGSETPGTSESATPES (16 aa) | Monomeric Streptavidin | 114 aa | Biotin | ~244 (MW) | dsDNA | [ |
| Cas9-Avidin/ ssDNA | Cas9 C-terminus | SGSETPGTSESATPES (16 aa) | Avidin | 152 aa | Biotin | ~244 (MW) | ssDNA | [ |
| sgRNA-S1m/ ssDNA | sgRNA | S1m* | Recombinant Streptavidin | 159 aa | Biotin | ~244 (MW) | ssDNA | [ |
| Cas9-PVC/ ssDNA | Cas9 C-terminus | H4-2 | Porcine Circovirus 2 (PCV) Rep | 109 aa | PCV.BS | 13 nt | ssDNA | [ |
| Cas9-VirD2/ ssDNA (Plant editing) | Cas9 | Undeclared | VirD2 | 455 aa | T-DNA.BS | 25 nt | ssDNA | [ |
| Cas9-THAP11/ dsDNA | Cas9 C-terminus | SGSETPGTSESATPES (16 aa) | THAP11 | 105 aa | THAP11.BS | 19 bp | dsDNA | [ |
| Cas9-N57/ dsDNA | Cas9 C-terminus | GGGGGSGGGGSGGGGSGGGGSLDPGGGGSG (30 aa) | N57 | 57 aa | N57.BS | 292 bp | dsDNA | [ |
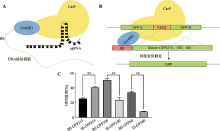
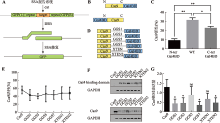
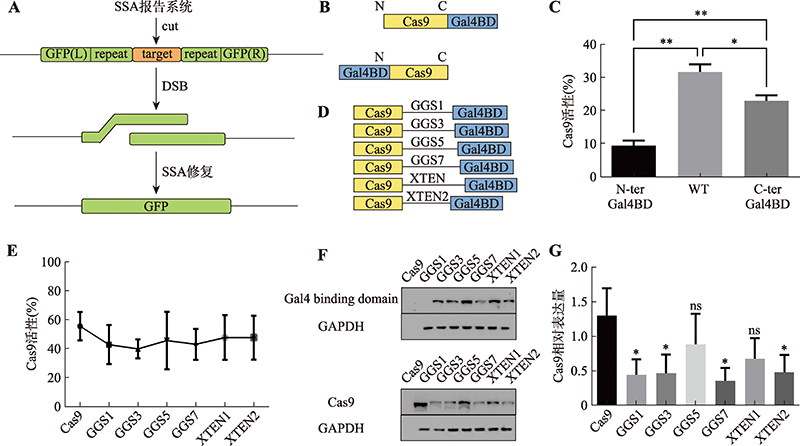
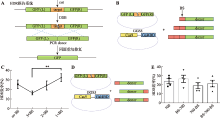
| Cas9-Gal4BD/ dsDNA | Cas9 C-terminus | 5×GGS (15 aa) | Gal4BD | 146 aa | Gal4BD.BS | 17 bp | dsDNA | This study |
| [1] |
Zhou SW, Yu HH, Zhao XE, Cai B, Ding Q, Huang Y, Li YX, Li Y, Niu YY, Lei AM, Kou QF, Huang XX, Petersen B, Ma BH, Chen YL, Wang XL. Generation of gene-edited sheep with a defined Booroola fecundity gene (FecB B) mutation in bone morphogenetic protein receptor type 1B (BMPR1B) via clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) 9. Reprod Fertil Dev, 2018, 30(12): 1616-1621.
doi: 10.1071/RD18086 |
| [2] |
Niu YY, Zhao XE, Zhou JK, Li Y, Huang Y, Cai B, Liu YT, Ding Q, Zhou SW, Zhao J, Zhou GX, Ma BH, Huang XX, Wang XL, Chen YL.Efficient generation of goats with defined point mutation (I397V) in GDF9 through CRISPR/Cas9. Reprod Fertil Dev, 2018, 30(2): 307-312.
doi: 10.1071/RD17068 |
| [3] |
Park KE, Kaucher AV, Powell A, Waqas MS, Sandmaier SES, Oatley MJ, Park CH, Tibary A, Donovan DM, Blomberg LA, Lillico SG, Whitelaw CBA, Mileham A, Telugu BP, Oatley JM. Generation of germline ablated male pigs by CRISPR/Cas9 editing of the NANOS2 gene. Sci Rep, 2017, 7: 40176.
doi: 10.1038/srep40176 |
| [4] |
Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell, 2014, 157(6): 1262-1278.
doi: 10.1016/j.cell.2014.05.010 |
| [5] |
Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kühn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol, 2015, 33(5): 543-548.
doi: 10.1038/nbt.3198 |
| [6] |
Decottignies A. Alternative end-joining mechanisms: a historical perspective. Front Genet, 2013, 4: 48.
doi: 10.3389/fgene.2013.00048 pmid: 23565119 |
| [7] |
Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc Natl Acad Sci USA, 2001, 98(15): 8403-10.
doi: 10.1073/pnas.111009698 |
| [8] |
Ruff P, Koh KD, Keskin H, Pai RB, Storici F. Aptamer-guided gene targeting in yeast and human cells. Nucleic Acids Res, 2014, 42(7): e61.
doi: 10.1093/nar/gku101 |
| [9] |
Savic N, Ringnalda FC, Lindsay H, Berk C, Bargsten K, Li YZ, Neri D, Robinson MD, Ciaudo C, Hall J, Jinek M, Schwank G. Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology- directed repair. eLife, 2018, 7: e33761.
doi: 10.7554/eLife.33761 |
| [10] |
Gu B, Posfai E, Rossant J. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat Biotechnol, 2018, 36(7): 632-637.
doi: 10.1038/nbt.4166 |
| [11] |
Ma M, Zhuang FF, Hu XB, Wang BL, Wen XZ, Ji JF, Xi JZJ. Efficient generation of mice carrying homozygous double-floxp alleles using the Cas9-Avidin/Biotin-donor DNA system. Cell Res, 2017, 27(4): 578-581.
doi: 10.1038/cr.2017.29 |
| [12] |
Carlson-Stevermer J, Abdeen AA, Kohlenberg L, Goedland M, Molugu K, Lou M, Saha K. Assembly of CRISPR ribonucleoproteins with biotinylated oligonucleotides via an RNA aptamer for precise gene editing. Nat Commun, 2017, 8(1): 1711.
doi: 10.1038/s41467-017-01875-9 pmid: 29167458 |
| [13] |
Aird EJ, Lovendahl KN, Martin AS, Harris RS, Gordon WR. Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun Biol, 2018, 1: 54.
doi: 10.1038/s42003-018-0054-2 |
| [14] |
Ali Z, Shami A, Sedeek K, Kamel R, Alhabsi A, Tehseen M, Hassan N, Butt H, Kababji A, Hamdan SM, Mahfouz MM. Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun Biol, 2020, 3(1): 44.
doi: 10.1038/s42003-020-0768-9 |
| [15] |
Li GL, Wang HQ, Zhang XW, Wu ZF, Yang HQ. A Cas9- transcription factor fusion protein enhances homology- directed repair efficiency. J Biol Chem, 2021, 296: 100525.
doi: 10.1016/j.jbc.2021.100525 |
| [16] |
Ma SF, Wang XL, Hu YF, Lv J, Liu CF, Liao KT, Guo XH, Wang D, Lin Y, Rong ZL. Enhancing site-specific DNA integration by a Cas9 nuclease fused with a DNA donor-binding domain. Nucleic Acids Res, 2020, 48(18): 10590-10601.
doi: 10.1093/nar/gkaa779 |
| [17] |
Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol, 2015, 33(5): 538-542.
doi: 10.1038/nbt.3190 pmid: 25798939 |
| [18] | Hu Z, Shi Z, Guo X, Jiang B, Wang G, Luo D, Chen Y, Zhu YS. Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR-Cas9 and ssODN in human cancer cells. Cell Biosci. 2018 19(8):12. |
| [19] |
Riesenberg S, Maricic T. Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat Commun, 2018, 9(1): 2164.
doi: 10.1038/s41467-018-04609-7 pmid: 29867139 |
| [20] |
Riesenberg S, Chintalapati M, Macak D, Kanis P, Maricic T, Pääbo S. Simultaneous precise editing of multiple genes in human cells. Nucleic Acids Res. 2019 47(19):e116.
doi: 10.1093/nar/gkz669 |
| [21] |
Robert F, Barbeau M, Éthier S, Dostie J, Pelletier J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med, 2015, 7(1): 93.
doi: 10.1186/s13073-015-0215-6 |
| [22] |
Li G, Liu D, Zhang X, Quan R, Zhong C, Mo J, Huang Y, Wang H, Ruan X, Xu Z, Zheng E, Gu T, Hong L, Li Z, Wu Z, Yang H. Suppressing Ku70/Ku80 expression elevates homology-directed repair efficiency in primary fibroblasts. Int J Biochem Cell Biol, 2018, 99: 154-160.
doi: 10.1016/j.biocel.2018.04.011 |
| [23] |
Nambiar TS, Billon P, Diedenhofen G, Hayward SB, Taglialatela A, Cai KH, Huang JW, Leuzzi G, Cuella-Martin R, Palacios A, Gupta A, Egli D, Ciccia A. Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat Commun, 2019, 10(1): 3395.
doi: 10.1038/s41467-019-11105-z pmid: 31363085 |
| [24] |
Paulsen BS, Mandal PK, Frock RL, Boyraz B, Yadav R, Upadhyayula S, Gutierrez-Martinez P, Ebina W, Fasth A, Kirchhausen T, Talkowski ME, Agarwal S, Alt FW, Rossi DJ. Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing. Nat Biomed Eng, 2017, 1(11): 878-888.
doi: 10.1038/s41551-017-0145-2 pmid: 31015609 |
| [25] |
Tran NT, Bashir S, Li X, Rossius J, Chu VT, Rajewsky K, Kühn R. Enhancement of precise gene editing by the association of Cas9 with homologous recombination Factors. Front Genet, 2019, 10: 365.
doi: 10.3389/fgene.2019.00365 |
| [26] |
Hackley CR. A novel set of Cas9 fusion proteins to stimulate homologous recombination: Cas9-HRs. CRISPR J, 2021, 4(2): 253-263.
doi: 10.1089/crispr.2020.0034 pmid: 33876961 |
| [27] |
Shao SM, Ren CH, Liu ZT, Bai YC, Chen ZL, Wei ZH, Wang X, Zhang ZY, Xu K.Enhancing CRISPR/Cas9- mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int J Biochem Cell Biol, 2017, 92: 43-52.
doi: 10.1016/j.biocel.2017.09.012 |
| [28] |
Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol, 2016, 34(3): 339-344.
doi: 10.1038/nbt.3481 |
| [29] |
Shy BR, Vykunta V, Ha A, Roth TL, Talbot A, Nguyen DN, Chen YY, Blaeschke F, Vedova S, Mamedov MR, Chung JY, Li H, Wolf J, Martin TG, Ye LM, Eyquem J, Esensten JH, Marson A. Hybrid ssDNA repair templates enable high yield genome engineering in primary cells for disease modeling and cell therapy manufacturing. bioRxiv, 2021, doi: 10.1101/2021.09.02.458799.
doi: 10.1101/2021.09.02.458799 |
| [30] |
Cruz-Becerra G, Kadonaga JT. Enhancement of homology-directed repair with chromatin donor templates in cells. eLife, 2020, 9: e55780.
doi: 10.7554/eLife.55780 |
| [31] |
Hirotsune S, Kiyonari H, Jin MY, Kumamoto K, Yoshida K, Shinohara M, Watanabe H, Wynshaw-Boris A, Matsuzaki F. Enhanced homologous recombination by the modulation of targeting vector ends. Sci Rep, 2020, 10(1): 2518.
doi: 10.1038/s41598-020-58893-9 pmid: 32054870 |
| [32] |
Liang X, Potter J, Kumar S, Ravinder N, Chesnut JD. Enhanced CRISPR/Cas9-mediated precise genome editing by improved design and delivery of gRNA, Cas9 nuclease, and donor DNA. J Biotechnol, 2017, 241: 136-146.
doi: 10.1016/j.jbiotec.2016.11.011 |
| [33] |
Nguyen DN, Roth TL, Li PJ, Chen PA, Apathy R, Mamedov MR, Vo LT, Tobin VR, Goodman D, Shifrut E, Bluestone JA, Puck JM, Szoka FC, Marson A. Polymer- stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat Biotechnol, 2020, 38(1): 44-49.
doi: 10.1038/s41587-019-0325-6 pmid: 31819258 |
| [34] |
Ling X, Xie B, Gao X, Chang L, Zheng W, Chen H, Huang Y, Tan L, Li M, Liu T. Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates. Sci Adv, 2020, 6(15): eaaz0051.
doi: 10.1126/sciadv.aaz0051 |
| [35] | Lomova A, Clark DN, Campo-Fernandez B, Flores- Bjurström C, Kaufman ML, Fitz-Gibbon S, Wang XY, Miyahira EY, Brown D, DeWitt MA, Corn JE, Hollis RP, Romero Z, Kohn DB. Improving gene editing outcomes in human hematopoietic stem and progenitor cells by temporal control of DNA repair. Stem Cells, 2019, 37(2): 284-294. |
| [36] |
Wienert B, Nguyen DN, Guenther A, Feng SJ, Locke MN, Wyman SK, Shin J, Kazane KR, Gregory GL, Carter MAM, Wright F, Conklin BR, Marson A, Richardson CD, Corn JE.Timed inhibition of CDC7 increases CRISPR- Cas9 mediated templated repair. Nat Commun, 2020, 11(1): 2109.
doi: 10.1038/s41467-019-13787-x |
| [37] |
Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu YW, Xu J, Chun N, Yuan W, Cheng T, Zhang XB. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol, 2017, 18(1): 35.
doi: 10.1186/s13059-017-1164-8 |
| [38] |
Gutschner T, Haemmerle M, Genovese G, Draetta GF, Chin L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep, 2016, 14(6): 1555-1566.
doi: S2211-1247(16)00040-1 pmid: 26854237 |
| [39] |
Matsumoto D, Tamamura H, Nomura W. A cell cycle- dependent CRISPR-Cas9 activation system based on an anti-CRISPR protein shows improved genome editing accuracy. Commun Biol, 2020, 3(1): 601-601.
doi: 10.1038/s42003-020-01340-2 pmid: 33097793 |
| [40] |
Li G, Zhang X, Zhong C, Mo J, Quan R, Yang J, Liu D, Li Z, Yang H, Wu Z. Small molecules enhance CRISPR/ Cas9-mediated homology-directed genome editing in primary cells. Sci Rep, 2017, 7(1): 8943.
doi: 10.1038/s41598-017-09306-x |
| [41] |
Takayama K, Igai K, Hagihara Y, Hashimoto R, Hanawa M, Sakuma T, Tachibana M, Sakurai F, Yamamoto T, Mizuguchi H. Highly efficient biallelic genome editing of human ES/iPS cells using a CRISPR/Cas9 or TALEN system. Nucleic Acids Res, 2017, 45(9): 5198-5207.
doi: 10.1093/nar/gkx130 pmid: 28334759 |
| [42] |
Li GL, Zhang XW, Wang HQ, Liu DW, Li ZC, Wu ZF, Yang HQ. Increasing CRISPR/Cas9-mediated homology- directed DNA repair by histone deacetylase inhibitors. Int J Biochem Cell Biol, 2020, 125: 105790.
doi: 10.1016/j.biocel.2020.105790 |
| [43] |
Keegan L, Gill G, Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science, 1986, 231(4739): 699-704.
pmid: 3080805 |
| [44] |
Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature, 1992, 356(6368): 408-414.
doi: 10.1038/356408a0 |
| [45] |
Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J, 1995, 9(9): 777-787.
pmid: 7601342 |
| [46] |
Xu K, Ren CH, Liu ZT, Zhang T, Zhang TT, Li D, Wang L, Yan Q, Guo LJ, Shen JC, Zhang ZY.Efficient genome engineering in eukaryotes using Cas9 from Streptococcus thermophilus. Cell Mol Life Sci, 2015, 72(2): 383-399.
doi: 10.1007/s00018-014-1679-z |
| [47] |
Liang SD, Marmorstein R, Harrison SC, Ptashne M. DNA sequence preferences of GAL4 and PPR1: how a subset of Zn2 Cys6 binuclear cluster proteins recognizes DNA. Mol Cell Biol, 1996, 16(7): 3773-3780.
doi: 10.1128/MCB.16.7.3773 pmid: 8668194 |
| [48] |
Bram RJ, Lue NF, Kornberg RD. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J, 1986, 5(3): 603-608.
doi: 10.1002/j.1460-2075.1986.tb04253.x pmid: 3011415 |
| [49] |
Selleck SB, Majors JE. In vivo DNA-binding properties of a yeast transcription activator protein. Mol Cell Biol, 1987, 7(9): 3260-3267.
doi: 10.1128/mcb.7.9.3260-3267.1987 pmid: 3313011 |
| [50] |
Yan NN, Sun YS, Fang YY, Deng JR, Mu L, Xu K, Mymryk JS, Zhang ZY. A universal surrogate reporter for efficient enrichment of CRISPR/Cas9-mediated homology- directed repair in mammalian cells. Mol Ther Nucleic Acids, 2020, 19: 775-789.
doi: 10.1016/j.omtn.2019.12.021 |
| [51] |
Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, Kurita M, Hishida T, Li M, Aizawa E, Guo SC, Chen S, Goebl A, Soligalla RD, Qu J, Jiang TS, Fu X, Jafari M, Esteban CR, Berggren WT, Lajara J, Nuñez-Delicado E, Guillen P, Campistol JM, Matsuzaki F, Liu GH, Magistretti P, Zhang K, Callaway EM, Zhang K, Belmonte JCI. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature, 2016, 540(7631): 144-149.
doi: 10.1038/nature20565 |
| [1] | Zhong Bian, Dongping Cao, Wenshu Zhuang, Shuwei Zhang, Qiaoquan Liu, Lin Zhang. Revelation of rice molecular design breeding: the blend of tradition and modernity [J]. Hereditas(Beijing), 2023, 45(9): 718-740. |
| [2] | Bingzheng Wang, Chao Zhang, Jiali Zhang, Jin Sun. Conditional editing of the Drosophila melanogaster genome using single transcripts expressing Cas9 and sgRNA [J]. Hereditas(Beijing), 2023, 45(7): 593-601. |
| [3] | Meizhen Liu, Liren Wang, Yongmei Li, Xueyun Ma, Honghui Han, Dali Li. Generation of genetically modified rat models via the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2023, 45(1): 78-87. |
| [4] | Chong Zhang, Zixuan Wei, Min Wang, Yaosheng Chen, Zuyong He. Editing MC1R in human melanoma cells by CRISPR/Cas9 and functional analysis [J]. Hereditas(Beijing), 2022, 44(7): 581-590. |
| [5] | Yao Liu, Xianhui Zhou, Shuhong Huang, Xiaolong Wang. Prime editing: a search and replace tool with versatile base changes [J]. Hereditas(Beijing), 2022, 44(11): 993-1008. |
| [6] | Yuting Han, Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni. The cutting edge of gene regulation approaches in model organism Drosophila [J]. Hereditas(Beijing), 2022, 44(1): 3-14. |
| [7] | Guangwu Yang, Yuan Tian. The F-box gene Ppa promotes lipid storage in Drosophila [J]. Hereditas(Beijing), 2021, 43(6): 615-622. |
| [8] | Dingwei Peng, Ruiqiang Li, Wu Zeng, Min Wang, Xuan Shi, Jianhua Zeng, Xiaohong Liu, Yaoshen Chen, Zuyong He. Editing the cystine knot motif of MSTN enhances muscle development of Liang Guang Small Spotted pigs [J]. Hereditas(Beijing), 2021, 43(3): 261-270. |
| [9] | Na Wang, Zhilian Jia, Qiang Wu. RFX5 regulates gene expression of the Pcdhα cluster [J]. Hereditas(Beijing), 2020, 42(8): 760-774. |
| [10] | Guoling Li, Shanxin Yang, Zhenfang Wu, Xianwei Zhang. Recent developments in enhancing the efficiency of CRISPR/Cas9- mediated knock-in in animals [J]. Hereditas(Beijing), 2020, 42(7): 641-656. |
| [11] | Yingnan Chen, Jing Lu. Application of CRISPR/Cas9 mediated gene editing in trees [J]. Hereditas(Beijing), 2020, 42(7): 657-668. |
| [12] | Siyuan Liu, Guoqiang Yi, Zhonglin Tang, Bin Chen. Progress on genome-wide CRISPR/Cas9 screening for functional genes and regulatory elements [J]. Hereditas(Beijing), 2020, 42(5): 435-443. |
| [13] | Lianchao Tang, Feng Gu. Next-generation CRISPR-Cas for genome editing: focusing on the Cas protein and PAM [J]. Hereditas(Beijing), 2020, 42(3): 236-249. |
| [14] | Junxia Cao, Youliang Wang, Zhengxu Wang. Advances in precise regulation of CRISPR/Cas9 gene editing technology [J]. Hereditas(Beijing), 2020, 42(12): 1168-1177. |
| [15] | Liwen Bao, Yiye Zhou, Fanyi Zeng. Advances in gene therapy for β-thalassemia and hemophilia based on the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2020, 42(10): 949-964. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||