Hereditas(Beijing) ›› 2025, Vol. 47 ›› Issue (9): 992-1006.doi: 10.16288/j.yczz.24-345
• Review • Previous Articles Next Articles
Developmental regulatory factors promote the efficiency of crop genetic transformation
Yingying Xie1( ), Kejian Wang2, Yuchun Rao1(
), Kejian Wang2, Yuchun Rao1( ), Yong Huang2(
), Yong Huang2( )
)
- 1. College of Life Sciences, Zhejiang Normal University, Jinhua 321004, China
2. National Key Laboratory of Rice Biology and Breeding, China National Rice Research Institute, Hangzhou 310006, China
-
Received:2024-12-05Revised:2025-03-12Online:2025-04-18Published:2025-04-18 -
Contact:Yuchun Rao, Yong Huang E-mail:1485637288@qq.com;ryc@zjnu.cn;huangyong@caas.cn -
Supported by:National Natural Science Foundation of China(32101721);Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences(CAAS-ASTIP-2021-CNRRI);Central Public-Interest Scientific Institution Basal Research Fund(CPSIBRF-CNRRI-202409)
Cite this article
Yingying Xie, Kejian Wang, Yuchun Rao, Yong Huang. Developmental regulatory factors promote the efficiency of crop genetic transformation[J]. Hereditas(Beijing), 2025, 47(9): 992-1006.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
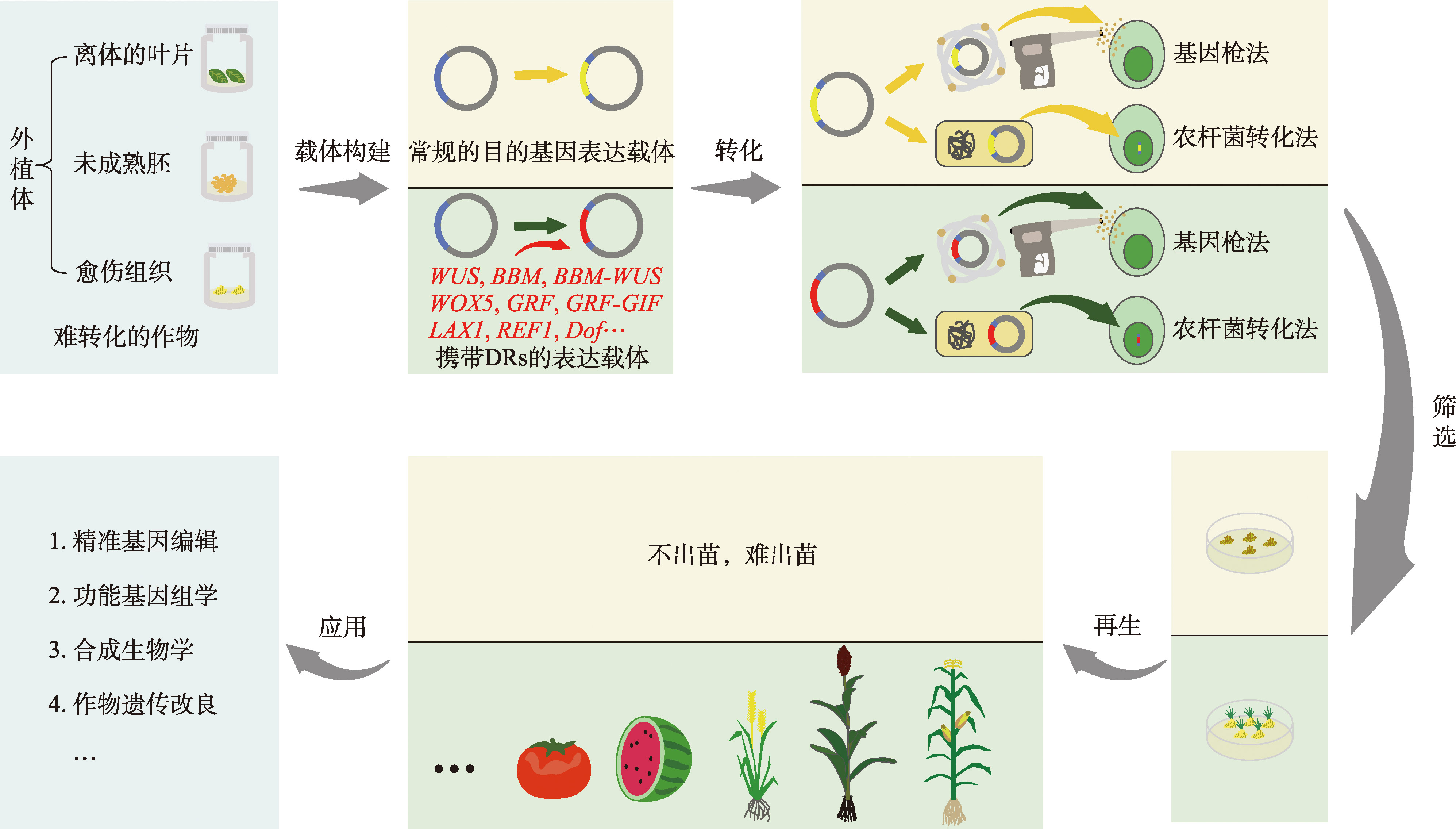
Table 1
Application of developmental regulatory factors to promote crop genetic transformation and regeneration"
| 转化与再生 | 发育调节因子 | 物种 | 遗传转化方法 | 提升效果 | 参考文献 |
|---|---|---|---|---|---|
| 提高转化效率 | ZmWus2 | 高粱 | 农杆菌转化法 | 转化效率最高可达到69.7% | [ |
| BrrWUSa | 芜菁 | 农杆菌转化法 | 再生效率提高至13% | [ | |
| MDBBM | 苹果 | 农杆菌转化法 | 转化效率最高提高至30% | [ | |
| ZmBBM-ZmWUS2 | 玉米 | 农杆菌转化法 | 转化效率最高提高至51.7% | [ | |
| ZmBBM-ZmWUS2 | 高粱 | 农杆菌转化法 | 愈伤组织转化频率从1.9%增加到18.3% | [ | |
| OsBbm-ZmWUS2 | 水稻 | 农杆菌转化法 | 愈伤组织转化频率从0%增加到43% | [ | |
| ZmBBM-ZmWUS2 | 甘蔗 | 农杆菌转化法 | 愈伤组织转化频率最高提高了8.85倍 | [ | |
| Babyboom-Wuschel | 小麦 | 农杆菌转化法 | 转化效率为常规方法的5~6倍 | [ | |
| HvBBM-HvWUS | 大麦 | 农杆菌转化法 | 再生频率从7.32%增加到24.8% | [ | |
| 克服基因型依赖性 | TaWOX5 | 小麦 | 农杆菌转化法 | 转化效率最高可达到97.8% | [ |
| AtGRF5 | 甜菜 | 农杆菌转化法 | 转化效率提高了6倍 | [ | |
| AtGRF5 | 西瓜 | 农杆菌转化法 | 转化效率从0.88%提高至24.73% | [ | |
| TaGRF4-GIF1 | 小麦 | 农杆菌转化法 | 平均再生效率提高了7.8倍 | [ | |
| TaGRF4-GIF1 | 黑小麦 | 农杆菌转化法 | 再生效率提高至10.7% | [ | |
| TaGRF4-GIF1 | 水稻 | 农杆菌转化法 | 再生效率提高了2.1倍 | [ | |
| ClGRF4-GIF1 | 西瓜 | 农杆菌转化法 | 转化效率提高了近9倍 | [ | |
| GmGRF3-GIF1 | 大豆 | 农杆菌转化法 | 平均转化效率提高了2.74倍 | [ | |
| TaGRF4-OsGIF1 | 高粱 | 农杆菌转化法 | 转化效率最高可达到38.28% | [ | |
| 增强再生能力 | TaLAX1 | 小麦 | 农杆菌转化法 | 转化频率提高至79.52% | [ |
| ZmBA1 | 玉米 | 农杆菌转化法 | 再生效率从10.59%提高到36.40% | [ | |
| GmLAX1 | 大豆 | 农杆菌转化法 | 再生频率从15.68%提高到26.18% | [ | |
| ZmWIND1 | 玉米 | 农杆菌转化法 | 转化效率最高提高至37.5% | [ | |
| REF1 | 番茄 | 农杆菌转化法 | 再生和转化效率分别提高了19倍和12倍 | [ | |
| GmREF1 | 大豆 | 农杆菌转化法 | 再生和转化效率分别提高了9倍和5倍 | [ | |
| TaREF1 | 小麦 | 农杆菌转化法 | 再生和转化效率分别提高了8倍和4倍 | [ | |
| ZmREF1 | 玉米 | 农杆菌转化法 | 再生和转化效率分别提高了6倍和4倍 | [ | |
| TaDOF5.6 | 小麦 | 农杆菌转化法 | 转化效率最高提高至50% | [ | |
| TaDOF3.4 | 小麦 | 农杆菌转化法 | 转化效率最高提高至55% | [ | |
| GhAGL15 | 棉花 | 农杆菌转化法 | 愈伤组织形成率从38.1%增加到65.2% | [ |
| [1] |
Altpeter F, Springer NM, Bartley LE, Blechl AE, Brutnell TP, Citovsky V, Conrad LJ, Gelvin SB, Jackson DP, Kausch AP, Lemaux PG, Medford JI, Orozco-Cárdenas ML, Tricoli DM, Van Eck J, Voytas DF, Walbot V, Wang K, Zhang ZJ, Stewart CN Jr. Advancing crop transformation in the era of genome editing. Plant Cell, 2016, 28(7): 1510-1520.
pmid: 27335450 |
| [2] |
Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev, 2003, 67(1): 16-37.
pmid: 12626681 |
| [3] |
Hiei Y, Komari T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc, 2008, 3(5): 824-834.
pmid: 18451790 |
| [4] |
Ishida Y, Tsunashima M, Hiei Y, Komari T. Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol Biol, 2015, 1223: 189-198.
pmid: 25300841 |
| [5] |
Frame BR, Shou HX, Chikwamba RK, Zhang ZY, Xiang CB, Fonger TM, Pegg SEK, Li BC, Nettleton DS, Pei DQ, Wang K. Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiol, 2002, 129(1): 13-22.
pmid: 12011333 |
| [6] | Sanford JC. Biolistic plant transformation. Physiol Plantarum, 1990, 79(1): 206-209. |
| [7] |
Gordon-Kamm B, Sardesai N, Arling M, Lowe K, Hoerster G, Betts S, Jones AT. Using morphogenic genes to improve recovery and regeneration of transgenic plants. Plants (Basel), 2019, 8(2): 38.
pmid: 30754699 |
| [8] |
Srinivasan C, Liu ZR, Heidmann I, Supena EDJ, Fukuoka H, Joosen R, Lambalk J, Angenent G, Scorza R, Custers JBM, Boutilier K. Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta, 2007, 225(2): 341-351.
pmid: 16924539 |
| [9] |
Debernardi JM, Tricoli DM, Ercoli MF, Hayta S, Ronald P, Palatnik JF, Dubcovsky J. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat Biotechnol, 2020, 38(11): 1274-1279.
pmid: 33046875 |
| [10] |
Nelson-Vasilchik K, Hague J, Mookkan M, Zhang ZYJ, Kausch A. Transformation of recalcitrant sorghum varieties facilitated by Baby boom and Wuschel2. Curr Protoc Plant Biol, 2018, 3(4): e20076.
pmid: 30369099 |
| [11] |
Guo ML, Ye JY, Gao DW, Xu N, Yang J. Agrobacterium-mediated horizontal gene transfer: mechanism, biotechnological application, potential risk and forestalling strategy. Biotechnol Adv, 2019, 37(1): 259-270.
pmid: 30579929 |
| [12] | Hiei Y, Komari T. Improved protocols for transformation of indica rice mediated by Agrobacterium tumefaciens. Plant Cell Tiss Organ Cult, 2006, 85: 271-283. |
| [13] |
Ishida Y, Hiei Y, Komari T. Agrobacterium-mediated transformation of maize. Nat Protoc, 2007, 2(7): 1614-1621.
pmid: 17585302 |
| [14] |
Yassitepe JECT, da Silva VCH, Hernandes-Lopes J, Dante RA, Gerhardt IR, Fernandes FR, da Silva PA, Vieira LR, Bonatti V, Arruda P. Maize transformation: from plant material to the release of genetically modified and edited varieties. Front Plant Sci, 2021, 12: 766702.
pmid: 34721493 |
| [15] | Jones HD. Wheat transformation: current technology and applications to grain development and composition. J Cereal Sci, 2005, 41(2): 137-147. |
| [16] |
Ozyigit II, Yucebilgili Kurtoglu K. Particle bombardment technology and its applications in plants. Mol Biol Rep, 2020, 47(12): 9831-9847.
pmid: 33222118 |
| [17] |
Dong OX, Ronald PC. Targeted DNA insertion in plants. Proc Natl Acad Sci USA, 2021, 118(22): e2004834117.
pmid: 34050013 |
| [18] |
Wang M, Sun RR, Zhang BH, Wang QL. Pollen tube pathway-mediated cotton transformation. Methods Mol Biol, 2019, 1902: 67-73.
pmid: 30543062 |
| [19] | Zhou GY, Weng J, Zeng YS, Huang JG, Qian SY, Liu GL. Introduction of exogenous DNA into cotton embryos. Methods Enzymol, 1983, 101: 433-481. |
| [20] | Luo ZX, Wu R. A simple method for the transformation of rice via the pollen-tube pathway. Plant Mol Biol Rep, 1989, 7: 69-77. |
| [21] |
Yang LY, Cui GM, Wang YX, Hao YS, Du JZ, Zhang HM, Wang CB, Zhang HH, Wu SB, Sun Y. Expression of foreign genes demonstrates the effectiveness of pollen- mediated transformation in Zea mays. Front Plant Sci, 2017, 8: 383.
pmid: 28377783 |
| [22] | Kanwal M, Gogoi N, Jones B, Bariana H, Bansal U, Ahmad N. Pollen: a potential explant for genetic transformation in wheat (Triticum aestivum L.). Agronomy, 2022, 12(9): 2009. |
| [23] |
Chen WS, Chiu CC, Liu HY, Lee TL, Cheng JT, Lin CC, Wu YJ, Chang HY. Gene transfer via pollen-tube pathway for anti-fusarium wilt in watermelon. Biochem Mol Biol Int, 1998, 46(6): 1201-1209.
pmid: 9891853 |
| [24] |
Showalter AM, Heuberger S, Tabashnik BE, Carrière Y, Coates B. A primer for using transgenic insecticidal cotton in developing countries. J Insect Sci, 2009, 9: 22.
pmid: 19613464 |
| [25] |
Huang C, Yang C, Yang HF, Gong YD, Li XM, Li LX, Li L, Liu X, Li XY. Systematic investigation and validation of peanut genetic transformation via the pollen tube injection method. Plant Methods, 2024, 20(1): 190.
pmid: 39702279 |
| [26] | Song XM, Gu YH, Qin GY. Application of a transformation method via the pollen-tube pathway in agriculture molecular breeding. Life Sci, 2007, 4(1): 77-79. |
| [27] |
Long Y, Yang Y, Pan GT, Shen YO. New insights into tissue culture plant-regeneration mechanisms. Front Plant Sci, 2022, 13: 926752.
pmid: 35845646 |
| [28] |
Wang BB, Zhu L, Zhao BB, Zhao YP, Xie YR, Zheng ZG, Li YY, Sun J, Wang HY. Development of a haploid- inducer mediated genome editing system for accelerating maize breeding. Mol Plant, 2019, 12(4): 597-602.
pmid: 30902686 |
| [29] |
Hayta S, Smedley MA, Clarke M, Forner M, Harwood WA. An efficient Agrobacterium-mediated transformation protocol for hexaploid and tetraploid wheat. Curr Protoc, 2021, 1(3): e58.
pmid: 33656289 |
| [30] |
Chen CL, Hu YX, Ikeuchi M, Jiao YL, Prasad K, Su YH, Xiao J, Xu L, Yang WB, Zhao Z, Zhou WK, Zhou Y, Gao J, Wang JW. Plant regeneration in the new era: from molecular mechanisms to biotechnology applications. Sci China Life Sci, 2024, 67(7): 1338-1367.
pmid: 38833085 |
| [31] |
Ikeuchi M, Ogawa Y, Iwase A, Sugimoto K. Plant regeneration: cellular origins and molecular mechanisms. Development, 2016, 143(9): 1442-1451.
pmid: 27143753 |
| [32] |
Laux T, Mayer KF, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development, 1996, 122(1): 87-96.
pmid: 8565856 |
| [33] |
Lopes FL, Galvan-Ampudia C, Landrein B. WUSCHEL in the shoot apical meristem: old player, new tricks. J Exp Bot, 2021, 72(5): 1527-1535.
pmid: 33332559 |
| [34] |
Che P, Wu E, Simon MK, Anand A, Lowe K, Gao HR, Sigmund AL, Yang MZ, Albertsen MC, Gordon-Kamm W, Jones TJ. Wuschel2 enables highly efficient CRISPR/ Cas-targeted genome editing during rapid de novo shoot regeneration in sorghum. Commun Biol, 2022, 5(1): 344.
pmid: 35410430 |
| [35] |
Bouchabké-Coussa O, Obellianne M, Linderme D, Montes E, Maia-Grondard A, Vilaine F, Pannetier C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep, 2013, 32(5): 675-686.
pmid: 23543366 |
| [36] |
Liu YY, Zhang L, Li C, Yang YQ, Duan YW, Yang YP, Sun XD. Establishment of Agrobacterium-mediated genetic transformation and application of CRISPR/Cas9 genome-editing system to Brassica rapa var. rapa. Plant Methods, 2022, 18(1): 98.
pmid: 35933391 |
| [37] |
Li MF, Wrobel-Marek J, Heidmann I, Horstman A, Chen BJ, Reis R, Angenent GC, Boutilier K. Auxin biosynthesis maintains embryo identity and growth during BABY BOOM-induced somatic embryogenesis. Plant Physiol, 2022, 188(2): 1095-1110.
pmid: 34865162 |
| [38] |
Chen JJ, Tomes S, Gleave AP, Hall W, Luo ZW, Xu J, Yao JL. Significant improvement of apple (Malus domestica Borkh.) transgenic plant production by pre-transformation with a Baby boom transcription factor. Hortic Res, 2022, 9: uhab014.
pmid: 35039859 |
| [39] |
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho MJ, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang LJ, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao ZM, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng PZ, Bidney D, Falco C, Register J, Zhao ZY, Xu DP, Jones T, Gordon-Kamm W. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell, 2016, 28(9): 1998-2015.
pmid: 27600536 |
| [40] |
Lowe K, La Rota M, Hoerster G, Hastings C, Wang N, Chamberlin M, Wu E, Jones T, Gordon-Kamm W. Rapid genotype "independent" Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell Dev Biol Plant, 2018, 54(3): 240-252.
pmid: 29780216 |
| [41] | Zhou ZR, Yang YW, Ai G, Zhao MM, Han BZ, Zhao CJ, Chen YQ, Zhang YW, Pan H, Lan CX, Li Q, Xu JT, Yan WH. Boosting transformation in wheat by BBM-WUS. bioRxiv, 2022, doi: 10.1101/2022.03.13.483388. |
| [42] |
Suo JQ, Zhou CL, Zeng ZH, Li XP, Bian HW, Wang JH, Zhu MY, Han N. Identification of regulatory factors promoting embryogenic callus formation in barley through transcriptome analysis. BMC Plant Biol, 2021, 21(1): 145.
pmid: 33740900 |
| [43] |
Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, Laux T. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature, 2007, 446(7137): 811-814.
pmid: 17429400 |
| [44] |
Wang K, Shi L, Liang XN, Zhao P, Wang WX, Liu JX, Chang YN, Hiei Y, Yanagihara C, Du LP, Ishida Y, Ye XG. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat Plants, 2022, 8(2): 110-117.
pmid: 35027699 |
| [45] |
Mcfarland FL, Collier R, Walter N, Martinell B, Kaeppler SM, Kaeppler HF. A key to totipotency: Wuschel-like homeobox 2a unlocks embryogenic culture response in maize (Zea mays L.). Plant Biotechnol J, 2023, 21(9): 1860-1872.
pmid: 37357571 |
| [46] |
Omidbakhshfard MA, Proost S, Fujikura U, Mueller- Roeber B. Growth-Regulating Factors (GRFs): a small transcription factor family with important functions in plant biology. Mol Plant, 2015, 8(7): 998-1010.
pmid: 25620770 |
| [47] |
Kim JH, Tsukaya H. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. J Exp Bot, 2015, 66(20): 6093-6107.
pmid: 26160584 |
| [48] |
Kong JX, Martin-Ortigosa S, Finer J, Orchard N, Gunadi A, Batts LA, Thakare D, Rush B, Schmitz O, Stuiver M, Olhoft P, Pacheco-Villalobos D. Overexpression of the transcription factor GROWTH-REGULATING FACTOR5 improves transformation of dicot and monocot species. Front Plant Sci, 2020, 11: 572319.
pmid: 33154762 |
| [49] |
Pan WB, Cheng ZT, Han ZG, Yang H, Zhang WG, Zhang HW. Efficient genetic transformation and CRISPR/Cas9- mediated genome editing of watermelon assisted by genes encoding developmental regulators. J Zhejiang Univ Sci B, 2022, 23(4): 339-344.
pmid: 35403388 |
| [50] |
Feng Q, Xiao L, He YZ, Liu M, Wang JF, Tian SJ, Zhang X, Yuan L. Highly efficient, genotype-independent transformation and gene editing in watermelon (Citrullus lanatus) using a chimeric ClGRF4-GIF1 gene. J Integr Plant Biol, 2021, 63(12): 2038-2042.
pmid: 34862751 |
| [51] |
Zhao Y, Cheng P, Liu Y, Liu CY, Hu ZB, Xin DW, Wu XX, Yang ML, Chen QS. A highly efficient soybean transformation system using GRF3-GIF1 chimeric protein. J Integr Plant Biol, 2024, 67(1): 3-6.
pmid: 39240004 |
| [52] |
Li JP, Pan WB, Zhang S, Ma GJ, Li AX, Zhang HW, Liu LJ. A rapid and highly efficient sorghum transformation strategy using GRF4-GIF1/ternary vector system. Plant J, 2024, 117(5): 1604-1613.
pmid: 38038993 |
| [53] | Matin MN, Kang SG. Genetic and phenotypic analysis of lax1-6, a mutant allele of LAX PANICLE1 in rice. J Plant Biol, 2012, 55(1): 50-63. |
| [54] |
Yu Y, Yu HX, Peng J, Yao WJ, Wang YP, Zhang FL, Wang SR, Zhao YJ, Zhao XY, Zhang XS, Su YH. Enhancing wheat regeneration and genetic transformation through overexpression of TaLAX1. Plant Commun, 2024, 5(5): 100738.
pmid: 37897039 |
| [55] |
Zhu XQ, Xu ZP, Wang GY, Cong YL, Yu L, Jia RY, Qin Y, Zhang GY, Li B, Yuan DJ, Tu LL, Yang XY, Lindsey K, Zhang XL, Jin SX. Single-cell resolution analysis reveals the preparation for reprogramming the fate of stem cell niche in cotton lateral meristem. Genome Biol, 2023, 24(1): 194.
pmid: 37626404 |
| [56] |
Braybrook SA, Harada JJ. LECs go crazy in embryo development. Trends Plant Sci, 2008, 13(12): 624-630.
pmid: 19010711 |
| [57] |
Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc Natl Acad Sci USA, 2008, 105(8): 3151-3156.
pmid: 18287041 |
| [58] |
Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell, 1998, 93(7): 1195-1205.
pmid: 9657152 |
| [59] |
Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA, 2001, 98(20): 11806-11811.
pmid: 11573014 |
| [60] |
Brand A, Quimbaya M, Tohme J, Chavarriaga-Aguirre P. Arabidopsis LEC1 and LEC2 orthologous genes are key regulators of somatic embryogenesis in Cassava. Front Plant Sci, 2019, 10: 673.
pmid: 31191582 |
| [61] |
Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, Ohme-Takagi M. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol, 2011, 21(6): 508-514.
pmid: 21396822 |
| [62] |
Ikeuchi M, Sugimoto K, Iwase A. Plant callus: mechanisms of induction and repression. Plant Cell, 2013, 25(9): 3159-3173.
pmid: 24076977 |
| [63] |
Iwase A, Mita K, Nonaka S, Ikeuchi M, Koizuka C, Ohnuma M, Ezura H, Imamura J, Sugimoto K. WIND1- based acquisition of regeneration competency in Arabidopsis and rapeseed. J Plant Res, 2015, 128(3): 389-397.
pmid: 25810222 |
| [64] | Jiang YL, Wei X, Zhu MR, Zhang XY, Jiang QP, Wang ZX, Cao YY, An XL, Wan XY. Developmental regulators in promoting genetic transformation efficiency in maize and other plants. Curr Plant Biol, 2024, 40: 100383. |
| [65] |
Yang WT, Zhai HW, Wu FM, Deng L, Chao Y, Meng XW, Chen Q, Liu CH, Bie XM, Sun CL, Yu Y, Zhang XF, Zhang XY, Chang ZQ, Xue M, Zhao YJ, Meng XB, Li BS, Zhang XS, Zhang DJ, Zhao XY, Gao CX, Li JY, Li CY. Peptide REF1 is a local wound signal promoting plant regeneration. Cell, 2024, 187(12): 3024-3038.e3014.
pmid: 38781969 |
| [66] |
Gupta S, Malviya N, Kushwaha H, Nasim J, Bisht NC, Singh VK, Yadav D. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta, 2015, 241(3): 549-562.
pmid: 25564353 |
| [67] |
Liu XM, Bie XM, Lin XL, Li ML, Wang HZ, Zhang XY, Yang YM, Zhang CY, Zhang XS, Xiao J. Uncovering the transcriptional regulatory network involved in boosting wheat regeneration and transformation. Nat Plants, 2023, 9(6): 908-925.
pmid: 37142750 |
| [68] |
Joshi S, Paul P, Hartman JM, Perry SE. AGL15 promotion of somatic embryogenesis: role and molecular mechanism. Front Plant Sci, 2022, 13: 861556.
pmid: 35419012 |
| [69] |
Wang HA, Caruso LV, Downie AB, Perry SE. The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell, 2004, 16(5): 1206-1219.
pmid: 15084721 |
| [70] |
Perry SE, Nichols KW, Fernandez DE. The MADS domain protein AGL15 localizes to the nucleus during early stages of seed development. Plant Cell, 1996, 8(11): 1977-1989.
pmid: 8953767 |
| [71] |
Thakare D, Tang W, Hill K, Perry SE. The MADS-domain transcriptional regulator AGAMOUS-LIKE15 promotes somatic embryo development in Arabidopsis and soybean. Plant Physiol, 2008, 146(4): 1663-1672.
pmid: 18305206 |
| [72] |
Yang ZR, Li CF, Wang Y, Zhang CJ, Wu ZX, Zhang XY, Liu CL, Li FG. GhAGL15s, preferentially expressed during somatic embryogenesis, promote embryogenic callus formation in cotton (Gossypium hirsutum L.). Mol Genet Genomics, 2014, 289(5): 873-883.
pmid: 24833045 |
| [73] |
Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt ED, Boutilier K, Grossniklaus U, de Vries SC. The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol, 2001, 127(3): 803-816.
pmid: 11706164 |
| [74] |
Schmidt ED, Guzzo F, Toonen MA, De Vries SC. A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development, 1997, 124(10): 2049-2062.
pmid: 9169851 |
| [75] | Liu ZJ, Zhao YP, Zeng LH, Zhang Y, Wang YM, Hua JP. Characterization of GhSERK2 and its expression associated with somatic embryogenesis and hormones level in upland cotton. J Integr Agric, 2018, 17(3): 517-529. |
| [76] |
Thomas C, Meyer D, Himber C, Steinmetz A. Spatial expression of a sunflower SERK gene during induction of somatic embryogenesis and shoot organogenesis. Plant Physiol Biochem, 2004, 42(1): 35-42.
pmid: 15061082 |
| [77] |
Nolan KE, Kurdyukov S, Rose RJ. Expression of the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) gene is associated with developmental change in the life cycle of the model legume Medicago truncatula. J Exp Bot, 2009, 60(6): 1759-1771.
pmid: 19305022 |
| [78] |
Singla B, Khurana JP, Khurana P. Characterization of three somatic embryogenesis receptor kinase genes from wheat, riticum aestivum. TPlant Cell Rep, 2008, 27(5): 833-843.
pmid: 18210118 |
| [79] |
Elhiti M, Tahir M, Gulden RH, Khamiss K, Stasolla C. Modulation of embryo-forming capacity in culture through the expression of Brassica genes involved in the regulation of the shoot apical meristem. J Exp Bot, 2010, 61(14): 4069-4085.
pmid: 20729480 |
| [80] |
Purwestri YA, Lee YS, Meehan C, Mose W, Susanto FA, Wijayanti P, Fauzia AN, Nuringtyas TR, Hussain N, Putra HL, Gutierrez-Marcos J. RWP-RK Domain 3 (OsRKD3) induces somatic embryogenesis in black rice. BMC Plant Biol, 2023, 23(1): 202.
pmid: 37076789 |
| [81] |
Waki T, Hiki T, Watanabe R, Hashimoto T, Nakajima K. The Arabidopsis RWP-RK protein RKD4 triggers gene expression and pattern formation in early embryogenesis. Curr Biol, 2011, 21(15): 1277-1281.
pmid: 21802301 |
| [82] |
Fan MZ, Xu CY, Xu K, Hu YX. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res, 2012, 22(7): 1169-1180.
pmid: 22508267 |
| [83] |
Wang YL, Yang XD, Wang WL, Wang Y, Chen XS, Wu H, Gao ZY, Xu HH, Liu TK, Li Y, Xiao D, Liu WS, Hou XL, Zhang CW. Efficient genetic transformation and gene editing of Chinese cabbage using Agrobacterium rhizogenes. Plant Physiol, 2024, 197(2): kiae543.
pmid: 39404111 |
| [84] |
Chen KL, Wang YP, Zhang R, Zhang HW, Gao CX. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol, 2019, 70: 667-697.
pmid: 30835493 |
| [85] |
Mu JY, Tan HL, Zheng Q, Fu FY, Liang Y, Zhang J, Yang XH, Wang T, Chong K, Wang XJ, Zuo JR. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol, 2008, 148(2): 1042-1054.
pmid: 18689444 |
| [86] |
Minh-Thu PT, Kim JS, Chae S, Jun KM, Lee GS, Kim DE, Cheong JJ, Song SI, Nahm BH, Kim YK. A WUSCHEL homeobox transcription factor, OsWOX13, enhances drought tolerance and triggers early flowering in rice. Mol Cells, 2018, 41(8): 781-798.
pmid: 30078233 |
| [1] | Jiatong Yan, Guanwei Chen, Qingmei Miao, Cheng Peng, Lei Yang, Xiaoyun Chen, Xiaoli Xu, Wei Wei, Junfeng Xu, Xiaofu Wang. Onsite rapid detection method for genetically modified maize and soybean based on recombinase polymerase amplification [J]. Hereditas(Beijing), 2025, 47(6): 694-707. |
| [2] | Rui He, Xiujuan Zheng, Ningning Wang, Xuying Li, Mingqi Li, Shijing Nian, Kewei Wang. Identification of PANoptosis-related lncRNAs in hepatocellular carcinoma based on bioinformatics and construction of a prognostic model [J]. Hereditas(Beijing), 2025, 47(4): 456-475. |
| [3] | Liu Jixiang, Lai Siting, Bai Jing, Xu Jin. Il34 rescues metronidazole-induced impairment of spinal cord regeneration in zebrafish central nervous system [J]. Hereditas(Beijing), 2024, 46(6): 478-489. |
| [4] | Gang Zhang, Lin Zhu, Haojie Nie, Yuguo Bao, Yunlong Cheng. Application and prospect of BSA method based on bibliometrics in crop breeding [J]. Hereditas(Beijing), 2024, 46(5): 360-372. |
| [5] | Xiaojun Yang, Zhenhan Huang, Wei Liu, Wenqing Zhang, Zhibin Huang. Identification and functional characterization of CD209 homologous genes in zebrafish [J]. Hereditas(Beijing), 2024, 46(11): 947-957. |
| [6] | Mingjiang Chen, Guifu Liu, Yeqing Xiao, Hong Yu, Jiayang Li. Breeding of ZhongKeFaZaoGeng1 by molecular design [J]. Hereditas(Beijing), 2023, 45(9): 829-834. |
| [7] | Qianwen Lv, Yongfang Yang. The biological functions of peptide signaling in plant and the advances on its utilization for crop improvement [J]. Hereditas(Beijing), 2023, 45(9): 813-828. |
| [8] | Liumei Jian, Yingjie Xiao, Jianbing Yan. De novo domestication: a new way for crop design and breeding [J]. Hereditas(Beijing), 2023, 45(9): 741-753. |
| [9] | Qingyu Sun, Yang Zhou, Lijuan Du, Mengke Zhang, Jiale Wang, Yuanyuan Ren, Fang Liu. Analysis between macrophage-related genes with prognosis and tumor microenvironment in non-small cell lung cancer [J]. Hereditas(Beijing), 2023, 45(8): 684-699. |
| [10] | Yifan Yu, Zhen OuYang, Juan Guo, Yujun Zhao, Luqi Huang. Progress on regulatory elements of plant plastid genetic engineering [J]. Hereditas(Beijing), 2023, 45(6): 501-513. |
| [11] | Xue Lv, Bangjie Li, Hanmei Xu. Research progress on high throughput discovery and functional verification of functional micropeptides [J]. Hereditas(Beijing), 2022, 44(6): 478-490. |
| [12] | Wang Ya'nan, Tao Xu, Wanpeng Wang, Qingzhu Zhang, Xie Li'nan. Role of epigenetic modifications in the development of crops essential traits [J]. Hereditas(Beijing), 2021, 43(9): 858-879. |
| [13] | Xiangying Chen, Mengwei Li, Ying Wang, Quan Chen, Hanmei Xu. Progress on sORF-encoded micropeptides [J]. Hereditas(Beijing), 2021, 43(8): 737-746. |
| [14] | Ruiying Qin, Pengcheng Wei. Prime editing creates a novel dimension of plant precise genome editing [J]. Hereditas(Beijing), 2020, 42(6): 519-523. |
| [15] | Xinping Yang,Yuan Yu,Cao Xu. De novo domestication to create new crops [J]. Hereditas(Beijing), 2019, 41(9): 827-835. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||