Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (6): 565-576.doi: 10.16288/j.yczz.19-330
• Research Article • Previous Articles Next Articles
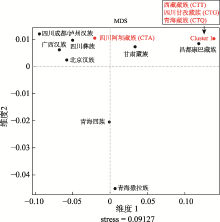
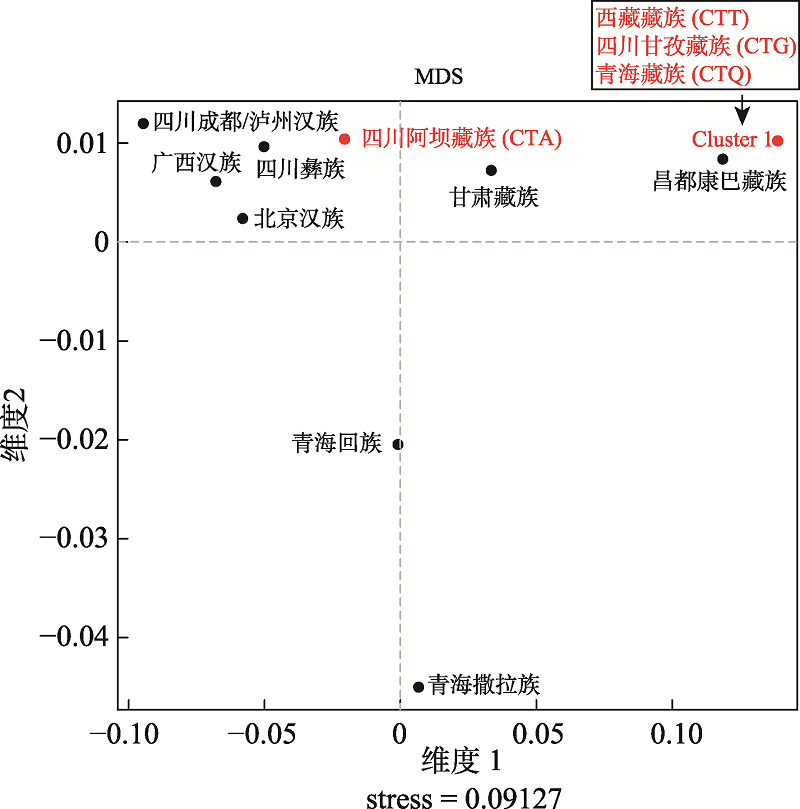
A genetic sub-structure study of the Tibetan population in Southwest China
Xiaojuan Wang1,2, Enfang Qian1, Yue Li2, Zhengyang Song1,2, Hui Zhao2, Hexin Xie2, Caixia Li1,2, Jiang Huang1, Li Jiang2
- 1. Institute of Forensic Medicine, Guizhou Medical University, Guiyang 550004, China
2. Institute of Forensic Science, Ministry of Public Security & Beijing Engineering Research Center of Crime Scene Evidence Examination & National Engineering Laboratory for Forensic Science, Beijing 100038, China;
-
Received:2019-12-26Revised:2020-04-14Online:2020-06-20Published:2020-04-17 -
Supported by:Supported by the National Natural Science Foundation of China No(81772027);the National Key R&D Program of China No(2017YFC0803501);the Fundamental Research Funds for Institute of Forensic Science No(2017JB025);the National Science and Technological Resources Platform No(YCZYPT[2017]01-3)
Cite this article
Xiaojuan Wang, Enfang Qian, Yue Li, Zhengyang Song, Hui Zhao, Hexin Xie, Caixia Li, Jiang Huang, Li Jiang. A genetic sub-structure study of the Tibetan population in Southwest China[J]. Hereditas(Beijing), 2020, 42(6): 565-576.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
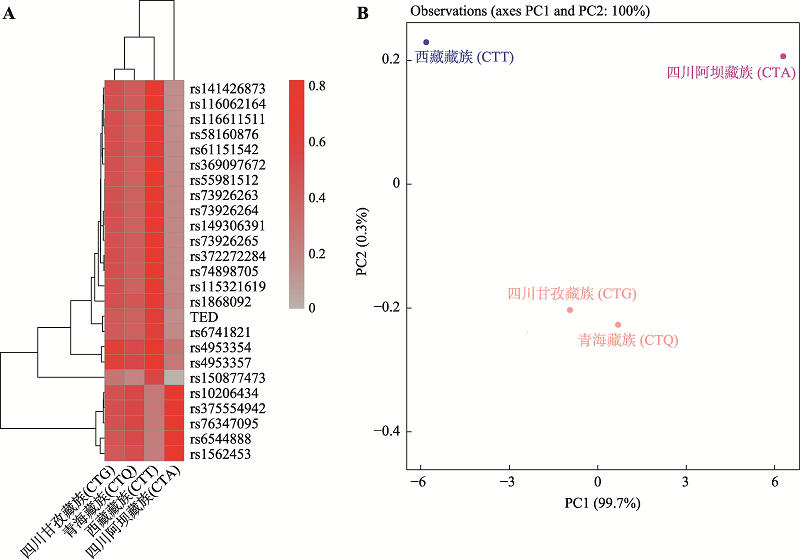
Table 1
Information of four Tibetan populations"
| 序号 | 群体 | 所属支系 | 英文全称 | 英文简称 | 检测样本量(例) | 海拔高度(m) |
|---|---|---|---|---|---|---|
| 1 | 西藏藏族 | 卫藏 | Chinese Tibetan in Tibetan Plateau | CTT | 89 | 3650 |
| 2 | 四川甘孜藏族 | 康巴 | Chinese Tibetan in Garze Sichuan, China | CTG | 88 | 3300 |
| 3 | 青海藏族 | 安多 | Chinese Tibetan in Qinghai, China | CTQ | 82 | 2420 |
| 4 | 四川阿坝藏族 | 嘉绒 | Chinese Tibetan in Aba Sichuan, China | CTA | 38 | 2130 |
Table 2
Y chromosome phylogenetic tree and haplogroup frequencies of four Tibetan populations"
| 单倍群谱系树 | 西藏藏族 (CTT) | 四川甘孜藏族 (CTG) | 青海藏族 (CTQ) | 四川阿坝藏族 (CTA) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | ||||||||||||||
| M168 | CT | 89 | 100 | 88 | 100 | 82 | 100 | 38 | 100 | ||||||||||||
| M130 | C | 2 | 2.25 | 2 | 2.27 | 2 | 2.44 | 1 | 2.63 | ||||||||||||
| M174 | D | 64 | 71.91 | 55 | 62.50 | 48 | 58.54 | 13 | 34.21 | ||||||||||||
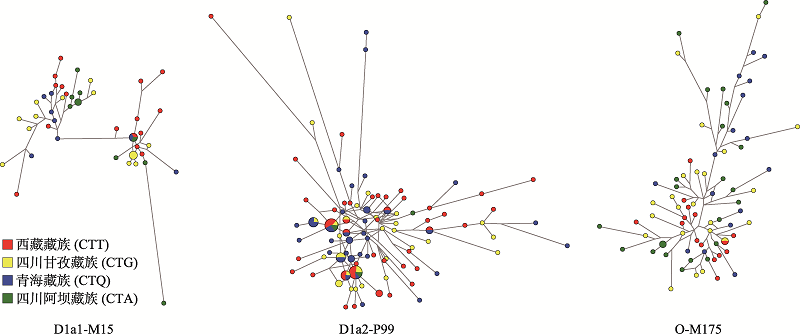
| M15 | D1a1 | 16 | 17.98 | 14 | 15.91 | 8 | 9.76 | 10 | 26.32 | ||||||||||||
| P99 | D1a2 | 48 | 53.93 | 41 | 46.59 | 40 | 48.78 | 3 | 7.89 | ||||||||||||
| M96 | E | 1 | 1.12 | 1 | 1.14 | 1 | 1.22 | ||||||||||||||
| M89 | F | 22 | 24.72 | 30 | 34.09 | 31 | 37.80 | 24 | 63.16 | ||||||||||||
| M201 | G | 1 | 1.22 | ||||||||||||||||||
| M304 | J | 4 | 4.88 | ||||||||||||||||||
| M9 | K | 22 | 24.72 | 30 | 34.09 | 26 | 31.71 | 24 | 63.16 | ||||||||||||
| M231 | N | 5 | 5.62 | 6 | 6.82 | 4 | 4.88 | ||||||||||||||
| M242 | Q | 2 | 2.25 | 1 | 1.22 | ||||||||||||||||
| M207 | R | 1 | 1.14 | 5 | 6.10 | 1 | 2.63 | ||||||||||||||
| M175 | O | 15 | 16.85 | 23 | 26.14 | 16 | 19.51 | 23 | 60.53 | ||||||||||||
| M122 | O2 | 15 | 16.85 | 21 | 23.86 | 12 | 14.63 | 20 | 52.63 | ||||||||||||
| M324 | O2a | 15 | 16.85 | 21 | 23.86 | 11 | 13.41 | 20 | 52.63 | ||||||||||||
| L127.1 | O2a1 | 1 | 1.12 | 3 | 3.41 | 5 | 6.10 | 5 | 13.16 | ||||||||||||
| P201 | O2a2 | 14 | 15.73 | 18 | 20.45 | 6 | 7.32 | 15 | 39.47 | ||||||||||||
| M117 | O2a2b1a1 | 12 | 13.48 | 10 | 11.36 | 4 | 4.88 | 8 | 21.05 | ||||||||||||
Table 3
Mitochondrial phylogenetic tree and haplogroup frequencies of each Tibetan population"
单倍群谱系树 | 西藏藏族 (CTT) | 四川甘孜藏族 (CTG) | 青海藏族 (CTQ) | 四川阿坝藏族 (CTA) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | ||||||||
| A1018G | L3 | 88 | 100.00 | 87 | 100.00 | 79 | 100.00 | 37 | 100.00 | ||||||
| G10400A | M | 59 | 67.05 | 54 | 62.07 | 48 | 60.76 | 21 | 56.76 | ||||||
| T4715C | M8 | 5 | 5.68 | 9 | 10.34 | 3 | 3.80 | 6 | 16.22 | ||||||
| A13263G | C | 3 | 3.41 | 8 | 9.20 | 2 | 2.53 | 3 | 8.11 | ||||||
| G4491A | M9 | 19 | 21.59 | 13 | 14.94 | 11 | 13.92 | 1 | 2.70 | ||||||
| A3394G | M9a'b | 16 | 18.18 | 9 | 10.34 | 8 | 10.13 | 1 | 2.70 | ||||||
| C7697T | M9a1a1c1b | 11 | 12.50 | 5 | 5.75 | 2 | 2.53 | ||||||||
| G14569A | M12'G | 13 | 14.77 | 10 | 11.49 | 7 | 8.86 | 4 | 10.81 | ||||||
| A4833G | G | 13 | 14.77 | 10 | 11.49 | 7 | 8.86 | 4 | 10.81 | ||||||
| T4561C | M62'68 | 1 | 1.27 | ||||||||||||
| G2735A | M62 | 1 | 1.27 | ||||||||||||
| C4883T | M80'D | 15 | 17.05 | 19 | 21.84 | 24 | 30.38 | 9 | 24.32 | ||||||
| C5178A | D | 15 | 17.05 | 19 | 21.84 | 24 | 30.38 | 9 | 24.32 | ||||||
| C3010T | D4 | 10 | 11.36 | 14 | 16.09 | 16 | 20.25 | 7 | 18.92 | ||||||
| C5262T | D4j1 | 4 | 4.55 | 3 | 3.45 | 2 | 2.53 | ||||||||
| C10873T | N | 29 | 32.95 | 33 | 37.93 | 31 | 39.24 | 16 | 43.24 | ||||||
| A663G | A | 10 | 11.36 | 5 | 5.75 | 10 | 12.66 | 3 | 8.11 | ||||||
| T12705 | R | 19 | 21.59 | 26 | 29.89 | 21 | 26.58 | 12 | 32.43 | ||||||
| C10310T | F | 11 | 12.50 | 14 | 16.09 | 10 | 12.66 | 3 | 8.11 | ||||||
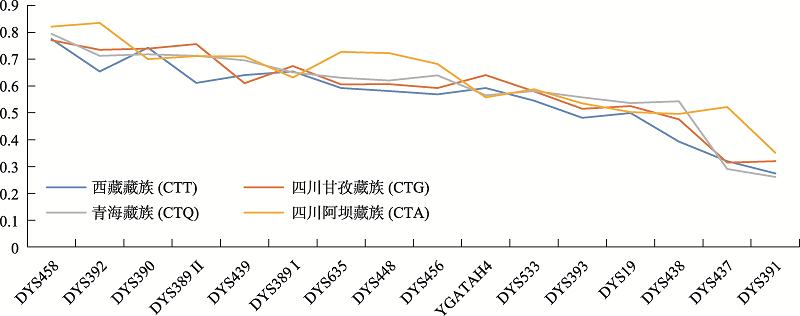
Supplementary Table 2
The weighting information of each Y-STR loci in Network analysis"
| 基因座 | 权重 | 突变率 | CTT | CTG | CTQ | CTA |
|---|---|---|---|---|---|---|
| DYS458 | 1 | 0.006 | 0.7750 | 0.7704 | 0.7943 | 0.8208 |
| DYS392 | 1 | 0.001 | 0.6545 | 0.7348 | 0.7124 | 0.8350 |
| DYS390 | 1 | 0.002 | 0.7421 | 0.7396 | 0.7185 | 0.6999 |
| DYS389 II | 2 | 0.004 | 0.6117 | 0.7565 | 0.7127 | 0.7112 |
| DYS439 | 2 | 0.005 | 0.6411 | 0.6108 | 0.6960 | 0.7098 |
| DYS389 I | 2 | 0.003 | 0.6540 | 0.6742 | 0.6501 | 0.6316 |
| DYS635 | 2 | 0.004 | 0.5924 | 0.6063 | 0.6308 | 0.7269 |
| DYS448 | 2 | 0.002 | 0.5812 | 0.6074 | 0.6208 | 0.7226 |
| DYS456 | 2 | 0.004 | 0.5687 | 0.5927 | 0.6399 | 0.6821 |
| GATA-H4 | 3 | 0.003 | 0.5927 | 0.6408 | 0.5652 | 0.5576 |
| DYS533 | 3 | 0.004 | 0.5457 | 0.5798 | 0.5815 | 0.5875 |
| DYS393 | 4 | 0.001 | 0.4814 | 0.5154 | 0.5580 | 0.5349 |
| DYS19 | 4 | 0.002 | 0.4990 | 0.5256 | 0.5363 | 0.5021 |
| DYS438 | 5 | 0.001 | 0.3933 | 0.4762 | 0.5434 | 0.4964 |
| DYS437 | 6 | 0.001 | 0.3202 | 0.3145 | 0.2906 | 0.5220 |
| DYS391 | 6 | 0.002 | 0.2743 | 0.3195 | 0.2611 | 0.3514 |
| [1] | Shi S . Tibetan regional characteristics and the related problems-and the Kham characteristic. J Qinghai Natl Inst(Soc Sci), 2015,41(1):207-208. |
| 石硕 . 藏族的地域特点及相关问题——兼论康区之特点. 青海民族大学学报(社会科学版), 2015,41(1):207-208. | |
| [2] | Dejizhuoga , The origin of Jirarong Tibetan. Tibet Stud, 2004, ( 2):51-56. |
| 德吉卓嘎 . 试论嘉绒藏族的族源. 西藏研究, 2004, ( 2):51-56. | |
| [3] | Majmundar AJ, Wong WJ, Simon MC . Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell, 2010,40(2):294-309. |
| [4] | Beall CM . Tibetan and Andean Patterns of adaptation to high-altitude hypoxia. Hum Biol, 2000,72(1):201-228. |
| [5] | Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZXP, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng HC, Liu T, He WM, Li K, Luo RB, Nie XF, Wu HL, Zhao MR, Cao HZ, Zou J, Shan Y, Li SZ, Yang Q, Asan, Ni PX, Tian G, Xu JM, Liu X, Jiang T, Wu RH, Zhou GY, Tang MF, Qin JJ, Wang T, Feng SJ, Li GH, Huasang, Luosang J, Wang W, Chen F, Wang YD, Zheng XG, Li Z, Bianba Z, Yang G, Wang XP, Tang SH, Gao GY, Chen Y, Luo Z, Gusang L, Cao Z, Zhang QH, Ouyang WH, Ren XL, Liang HQ, Zheng HS, Huang YB, Li JX, Bolund L, Kristiansen K, Li YR, Zhang Y, Zhang XQ, Li RQ, Li SG, Yang HM, Nielsen R, Wang J, Wang J. Sequencing of 50 human exomes reveals adaptation to high altitude. Science, 2010,329(5987):75-78. |
| [6] | Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, Prchal JT, Ge R . Genetic evidence for high-altitude adaptation in Tibet. Science, 2010,329(5987):72-75. |
| [7] | Huerta-Sánchez E, Jin X, Asan, Bianba Z, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, Ni P, Wang B, Ou X, Huasang, Luosang J, Cuo ZX, Li K, Gao G, Yin Y, Wang W, Zhang X, Xu X, Yang H, Li Y, Wang J, Wang J, Nielsen R. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature, 2014,512(7513):194-197. |
| [8] | Jiang L, Peng JX, Huang MS, Liu J, Wang L, Ma Q, Zhao H, Yang X, Ji AQ, Li CX . Differentiation analysis for estimating individual ancestry from the Tibetan Plateau by an archaic altitude adaptation EPAS1 haplotype among East Asian populations. Int J Legal Med, 2018,132(50):1527-1535. |
| [9] | Xiang K, Ouzhuluobu, Peng Y, Yang Z, Zhang X, Cui C, Zhang H, Li M, Zhang Y, Bianba, Gonggalanzi, Basang, Ciwangsangbu, Wu T, Chen H, Shi H, Qi X, Su B,. Identification of a Tibetan-specific mutation in the hypoxic gene EGLN1 and its contribution to high-altitude adaptation. Mol Biol Evol, 2013,30(8):1889-1898. |
| [10] | Wen B . Y chromosome, mtDNA polymorphism and genetic structure of East Asian population [Dissertation]. Shanghai: Fudan Univ, 2004. |
| 文波 . Y染色体、mtDNA多态性与东亚人群的遗传结构[学位论文]. 复旦大学, 2004. | |
| [11] | Hammer MF, Karafet TM, Park H, Omoto K, Harihara S, Stoneking M, Horai S . Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J Hum Genet, 2006,51(1):47-58. |
| [12] | Thangaraj K, Singh L, Reddy AG, Rao VR, Sehgal SC, Underhill PA, Pierson M, Frame IG, Hagelberg E . Genetic affinities of the Andaman Islanders, a vanishing human population. Curr Biol, 2003,13(2):86-93. |
| [13] | Karafet T, Xu L, Du R, Wang W, Feng S, Wells RS, Redd AJ, Zegura SL, Hammer MF . Paternal population history of East Asia: sources, patterns, and microevolutionary processes. Am J Hum Genet, 2001,69(3):615-628. |
| [14] | Bhandari S, Zhang XM, Cui CY, Bianba, Liao SY, Peng Y, Zhang H, Xiang K, Shi H, Ouzhuluobu, Baimakongzhu, Gonggalanzi, Liu SM, Gengdeng, Wu TY, Qi XB, Su B. Genetic evidence of a recent Tibetan ancestry to Sherpas in the Himalayan region. Sci Rep, 2015,5:16249. |
| [15] | Huang MS, Ma Q, Wang L, Ma X, Li CX, Jiang L . The study of Tibetan ancestry informative SNPs on high-altitude adaptive genes. Chin J Foren Med, 2017,32(6):588-599. |
| 黄美莎, 马泉, 王玲, 马新, 李彩霞, 江丽 . 高原适应基因中藏族祖先信息位点的研究. 中国法医学杂志, 2017,32(6):588-599. | |
| [16] | Liu J, Li S, Jiang L, Zhao L, Zhao WT, Feng L, Liu HB, Ji AQ, Li CX . DNA ancestry analyzer: an automatic program for ancestry inference of unknown individuals. Life Sci Res, 2018,22(1):3-7. |
| 刘京, 李盛, 江丽, 赵蕾, 赵雯婷, 丰蕾, 刘海渤, 季安全, 李彩霞 . 对于未知来源个体进行族群推断的自动分析系统. 生命科学研究, 2018,22(1):3-7. | |
| [17] | Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF . New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res, 2008,18(5):830-838. |
| [18] | Bandelt HJ, Forster P, Röhl A . Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol, 1999,16(1):37-48. |
| [19] | Wang H, Mao J, Xia Y, Bai XG, Zhu WQ, Peng D, Liang WB . Genetic polymorphisms of 17 Y-chromosomal STRs in the Chengdu Han population of China. Int J Legal Med, 2017,131(4):967-968. |
| [20] | Hidding M, Schmitt C . Haplotype frequencies and population data of nine Y-chromosomal STR polymorphisms in a German and a Chinese population. Forensic Sci Int, 2000,113(1-3):47-53. |
| [21] | Shi MS, Li YB, Tang JP, Zhang HJ, Hou YP . Southwest China Han Population data for nine Y-STR loci by multiplex polymerase chain reaction. J Forensic Sci, 2007,52(1):228-230. |
| [22] | Fan GY, An YR, Peng CX, Deng JL, Pan LP, Ye Y . Forensic and phylogenetic analyses among three Yi populations in Southwest China with 27 Y chromosomal STR loci. Int J Legal Med, 2018,133(3):795-797. |
| [23] | He GL, Wang Z, Su YD, Zou X, Wang MG, Chen X, Gao B, Liu J, Wang SY, Hou YP . Genetic structure and forensic characteristics of Tibeto-Burman-speaking Ü-Tsang and Kham Tibetan Highlanders revealed by 27 Y-chromosomal STRs. Sci Rep, 2019,9(1):7739. |
| [24] | Zhao Q, Bian Y, Zhang S, Zhu R, Zhou W, Gao Y, Li C . Population genetics study using 26 Y-chromosomal STR loci in the Hui ethnic group in China. Forensic Sci Int Genet, 2017,28:e26-e27. |
| [25] | Xie MK, Song F, Li JN, Lang M, Luo HB, Wang Z, Wu J, Li CZ, Tian CC, Wang WZ, Ma H, Song Z, Fan YJ, Hou YP . Genetic substructure and forensic characteristics of Chinese Hui populations using 157 Y-SNPs and 27 Y-STRs. Forensic Sci Int Genet, 2019,41:11-18. |
| [26] | Nothnagel M, Fan GY, Guo F, He YF, Hou YP, Hu SP, Huang J, Jiang XH, Kim W, Kim K, Li CT, Li H, Li LM, Li SL, Li Z, Liang WB, Liu C, Lu D, Luo HB, Nie SJ, Shi MS, Sun HY, Tang JP, Wang L, Wang CC, Wang D, Wen SQ, Wu HY, Wu WW, Xing JX, Yan JW, Yan S, Yao HB, Ye Y, Yun LB, Zeng ZS, Zha L, Zhang SH, Zheng XF, Willuweit S, Roewer L . Revisiting the male genetic landscape of China: a multi-center study of almost 38,000 Y-STR haplotypes. Hum Genet, 2017,136(5):485-497. |
| [27] | Lang M, Liu H, Song F, Qiao XH, Ye Y, Ren H, Li JN, Huang J, Xie MK, Chen SJ, Song MY, Zhang YF, Qian XQ, Yuan TX, Wang Z, Liu YM, Wang MG, Liu YC, Liu J, Hou YP . Forensic characteristics and genetic analysis of both 27 Y-STRs and 143 Y-SNPs in Eastern Han Chinese population. Forensic Sci Int Genet, 2019,42:e13-e20. |
| [28] | Kwak KD, Jin HJ, Shin DJ, Kim JM, Roewer L, Krawczak M, Tyler-Smith C, Kim W . Y-chromosomal STR haplotypes and their applications to forensic and population studies in east Asia. Int J Legal Med, 2005,119(4):195-201. |
| [29] | Bigham AW, Lee FS . Human high-altitude adaptation: forward genetics meets the HIF pathway. Genes Dev, 2014,28(20):2189-2204. |
| [30] | Peng Y, Yang ZH, Zhang H, Cui CY, Qi XB, Luo XJ, Tao X, Wu TY, Ouzhuluobu, Basang, Ciwangsangbu, Danzengduojie, Chen H, Shi H, Su B. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol, 2011,28(2):1075-1081. |
| [31] | Ke JK . A study of polymorphism sites of HIF-1α and HIF-2α gene in three Tibetan groups of different altitude [Dissertation]. Peking Union Med Coll, 2010. |
| 柯金坤 . 不同海拔三个藏族人群HIF-1α基因与HIF-2α基因多态位点的比较研究[学位论文]. 北京协和医学院, 2010. | |
| [32] | Deng L, Zhang C, Yuan K, Gao Y, Pan YW, Ge XL, He YX, Yuan Y, Lu Y, Zhang XX, Chen H, Lou HY, Wang XJ, Lu DS, Liu JJ, Tian L, Feng QD, Khan A, Yang YJ, Jin ZB, Yang J, Lu F, Qu J, Kang LL, Su B, Xu SH . Prioritizing natural-selection signals from the deep-sequencing genomic data suggests multi-variant adaptation in Tibetan highlanders. Natl Sci Rev, 2019,6(6):1201-1222. |
| [33] | Ouzhuluobu, He YX, Lou HY, Cui CY, Deng L, Gao Y, Zheng WS, Guo YB, Wang XJ, Ning ZL, Li J, Li B, Bai CJ, Baimakangzhuo, Gonggalanzi, Dejiquzong, Bianba, Duojizhuoma Liu SM, Wu TY, Xu SH, Qi XB, Su B,. De novo assembly of a Tibetan genome and identification of novel structural variants associated with high-altitude adaptation. Natl Sci Rev, 2019,7(2):391-402. |
| [34] | Zhang Q, Ping J, Zhang HX, Kang B, Li YF, Zhou GQ . Genetic association of MKL1 gene polymorphisms with the high-altitude adaptation. Hereditas(Beijing), 2019,41(7):634-643. |
| 张晴, 平杰, 张昊翔, 康波, 李元丰, 周钢桥 . MKL1基因多态性与高原环境适应性的遗传关联研究. 遗传, 2019,41(7):634-643. | |
| [35] | Wang CC, Li H . Inferring human history in East Asia from Y chromosomes. Investig Genet, 2013,4(1):11. |
| [36] | Qi XB, Cui CY, Peng Y, Zhang XM, Yang ZH, Zhong H, Zhang H, Xiang K, Cao XY, Wang Y, Ouzhuluobu, Basang, Ciwangsangbu, Bianba, Gonggalanzi, Wu TY, Chen H, Shi H, Su B. Genetic evidence of paleolithic colonization and neolithic expansion of modern humans on the Tibetan Plateau. Mol Biol Evol, 2013,30(8):1761-1778. |
| [37] | Wen B, Xie X, Gao S, Li H, Shi H, Song X, Qian T, Xiao C, Jin J, Su B, Lu D, Chakraborty R, Jin L . Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. Am J Hum Genet, 2004,74(5):856-865. |
| [38] | Cavalli-Sforza LL, Feldman MW . The application of molecular genetic approaches to the study of human evolution. Nat Genet, 2003,33(Suppl.):266-275. |
| [39] | Chu X, Shan KR, Wen B, Qi XL, Li Y, Wu CX, Liu X, Zhao Y, Ren XL, Jin L . Analysis of polymorphisms in Y-DNA haplotypes and mtDNA haplogroups in Yao ethnic group from Guizhou. Hereditas(Beijing), 2006,28(2):153-158. |
| 褚迅, 单可人, 文波, 齐晓岚, 李毅, 吴昌学, 刘烜, 赵艳, 任锡麟, 金力 . 贵州瑶族3支系Y-DNA及线粒体DNA序列多态性分析. 遗传, 2006,28(2):153-158. | |
| [40] | Wang XQ, Wang CC, Deng QY, Li H. Genetic analysis of Y chromosome and mitochondrial DNA poly-morphism of Mulam ethnic group in Guangxi, China. Hereditas (Beijing), 2013,35(2):168-174. |
| 王晓庆, 王传超, 邓琼英, 李辉 . 广西仫佬族Y染色体和mtDNA的遗传结构分析. 遗传, 2013,35(2):168-174. | |
| [41] | Gu ML, Wang YJ, Shi L, Jiang F, Qiu MJ, Lin KQ, Tao YF, Shi L, Huang XQ, Liu B, Chu JY . Comparative analysis of the complete mitochondrial genome between Tibetan and Han population. Chin J Med Genet, 2008,25(4):382-386. |
| 顾明亮, 汪业军, 史磊, 姜枫, 邱梦洁, 林克勤, 陶玉芬, 史荔, 黄小琴, 刘斌, 褚嘉佑 . 藏汉民族线粒体基因组全序列的比较研究. 中华医学遗传学杂志, 2008,25(4):382-386. | |
| [42] | Zhao M, Kong QP, Wang HW, Peng MS, Xie XD, Wang WZ, Jia yang, Duan JG, Cai MC, Zhao SN, Cidanping cuo, Tu YQ, Wu SF, Yao YG, Bandelt HJ, Zhang YP. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc Natl Acad Sci USA, 2009,106(50):21230-21235. |
| [43] | Qin ZD, Yang YJ, Kang LL, Yan S, Cho K, Cai XY, Lu Y, Zheng HX, Zhu DC, Fei DM, Li SL, Jin L, Li H . A mitochondrial revelation of early human migrations to the Tibetan Plateau before and after the last glacial maximum. Am J Phys Anthropol, 2010,143(4):555-569. |
| [44] | Peng MS, Palanichamy MG, Yao YG, Mitra B, Cheng YT, Zhao M, Liu J, Wang HW, Pan H, Wang WZ, Zhang AM, Zhang W, Wang D, Zou Y, Yang Y, Chaudhuri TK, Kong QP, Zhang YP . Inland post-glacial dispersal in East Asia revealed by mitochondrial haplogroup M9a'b. BMC Biol, 2011,9:2. |
| [45] | Soares P, Trejaut JA, Loo JH, Hill C, Mormina M, Lee CL, Chen YM, Hudjashov G, Forster P, Macaulay V, Bulbeck D, Oppenheimer S, Lin M, Richards MB . Climate change and postglacial human dispersals in southeast Asia. Mol Biol Evol, 2008,25(6):1209-1218. |
| [46] | Meng M . Exploration of the relationship of Yi and Qiang in the Southwest ethnic groups of Han dynasty. Hist Res, 1985,1:11-32. |
| 蒙默 . 试论汉代西南民族中的“夷”与“羌”. 历史研究, 1985,1:11-32. | |
| [47] | Zheng D, Zhang QS, Wu SH . Mountain Geoecology and Sustainable Development of the Tibetan Plateau. Geojournal Library, 2000,57(2):203-204. |
| [48] | Wang CM . The changes of population size and structure in Garze prefecture based on the census data. J Sichuan Univ Natl, 2013,22(3):58-67. |
| 王长明 . 基于人口普查数据的甘孜州人口规模与结构变化. 四川民族学院学报, 2013,22(3):58-67. | |
| [49] | Li Q . The relationship between Jiarong, Jialiangyi, Ranmang and Ge populations - and explore the origin of Jiarong Tibetan. J Sichuan Univ Natl, 2010,19(4):1-6. |
| 李青 . 试论嘉绒、嘉良夷、冉駹与戈人的关系——兼论嘉绒藏族的族源. 四川民族学院学报, 2010,19(4):1-6. | |
| [50] | Xie L . On the cultural reform of the Han-Tibetan intermarriage and its evolution. J Neijiang Norm Univ, 2006,21(1):147-149. |
| 谢蕾 . 藏汉通婚的文化整合及演变. 内江师范学院学报, 2006,21(1):147-149. | |
| [51] | Yan MS. History of Marriage in Ancient China. Guizhou: Guizhou Ethnic Publishing House, 2003. |
| 阎明恕 . 中国古代和亲史. 贵州: 贵州民族出版社, 2003. |
| [1] | Fei Wang, Meng Wang, Xinghua Zhang, Keli Yu, Lianbin Zheng, Yajun Yang. Genetic substructure analysis of three isolated populations in southwest China [J]. Hereditas(Beijing), 2022, 44(5): 424-431. |
| [2] | Zilong Wen, Yiqiang Zhao. Progress on animal domestication under population genetics [J]. Hereditas(Beijing), 2021, 43(3): 226-239. |
| [3] | Xi Li, Haoyu Wang, Yueyan Cao, Qiang Zhu, Panyin Shu, Tingyun Hou, Yuting Wang, Ji Zhang. Forensic genomics research on microhaplotypes [J]. Hereditas(Beijing), 2021, 43(10): 962-971. |
| [4] | Xiuquan Zhang,Jian Wang,Fu Xiong,Weibiao Lv,Yuanqing Zhou,Shaomin Yang,Yuting Zhang,Xiaoyan Tian,Wei Lian,Xiangmin Xu. Congenital ectrodactyly caused by chromosome 10q24.31 duplication and its pathogenetic analysis [J]. Hereditas(Beijing), 2019, 41(8): 716-724. |
| [5] | Jia Chen,Mingyue Shu,Jin Li,Aisi Fu,Fan Yang,Zou Wang,Yirong Li,Zixin Deng,Tiangang Liu. The third-generation sequencing combined with targeted capture technology for high-resolution HLA typing and MHC region haplotype identification [J]. Hereditas(Beijing), 2019, 41(4): 337-348. |
| [6] | Jian Hu,Yiren Zhou,Jialin Ding,Zhiyuan Wang,Ling Liu,Yekai Wang,Huiling Lou,Shouyi Qiao,Yanhua Wu. Simplification of genotyping techniques of the ABO blood type experiment and exploration of population genetics [J]. Hereditas(Beijing), 2017, 39(5): 423-429. |
| [7] | Feng Gao, Haipeng Li. Application of computer simulators in population genetics [J]. Hereditas(Beijing), 2016, 38(8): 707-717. |
| [8] | Yunsheng Wang. Research progress of plant population genomics based on high-throughput sequencing [J]. Hereditas(Beijing), 2016, 38(8): 688-699. |
| [9] | Juan Liu, Yan Sun, Qiang Jiang, Chunhong Yang, Jinming Huang, Jianbin Li, Minghai Hou, Jifeng Zhong, Changfa Wang, Baoshen Liu. Functional haplotypes of INCENP affect promoter activity and bovine semen quality [J]. HEREDITAS(Beijing), 2016, 38(1): 62-71. |
| [10] | Meifen Xu, Yiqun He, Junwei Geng, Yanzi Meng, Han Yu, Zhi Lin, Suxue Shi, Ling Xue, Zhongqiu Lu, Minxin Guan. The mitochondrial tRNAMet/tRNAGlnA4401G and tRNACysG5821A mutations may be associated with hypertension in two Han Chinese families [J]. HEREDITAS, 2014, 36(2): 127-134. |
| [11] | LI Qian, LIU Shu-Yuan, LIN Ke-Qin, SUN Hao, YU Liang, HUANG Xiao-Qin, CHU Jia-You, YANG Zhao-Qing. Association between six single nucleotide polymorphisms of EGLN1 gene and adaptation to high-altitude hypoxia [J]. HEREDITAS, 2013, 35(8): 992-998. |
| [12] | RUAN Qing-Wei YU Zhuo-Wei BAO Zhi-Jun MA Yong-Xing. The relationship between the polymorphism of immunity genes and both aging and age-related diseases [J]. HEREDITAS, 2013, 35(7): 813-822. |
| [13] | LI Duo CHAI Zhi-Xin JI Qiu-Mei ZHANG Cheng-Fu XIN Jin-Wei ZHONG Jin-Cheng. Genetic diversity of DNA microsatellite for Tibetan Yak [J]. HEREDITAS, 2013, 35(2): 175-184. |
| [14] | BAI Ru-Feng, YANG Li-Hai, YUAN Li, LIANG Quan-Zeng, LU Di, YANG Xue, SHI Mei-Sen. Polymorphism of 17 Y-STR loci in She ethnic population in Fujian and genetic relationship with 11 populations [J]. HEREDITAS, 2012, 34(8): 1020-1030. |
| [15] | SHU Wei, LIN You-Kun, HUA Rong, LUO Yan-Yan, FANG Lin, XU Shu-Ru, HE Na, MA Jun, HU Qi-Ping, LI Xiao-Long, YUAN Zhi-Gang. Haplotype analysis for mucocutaneous venous malformations in a Chinese Han ethnic family [J]. HEREDITAS, 2012, 34(4): 431-436. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||