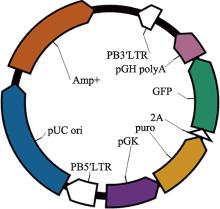
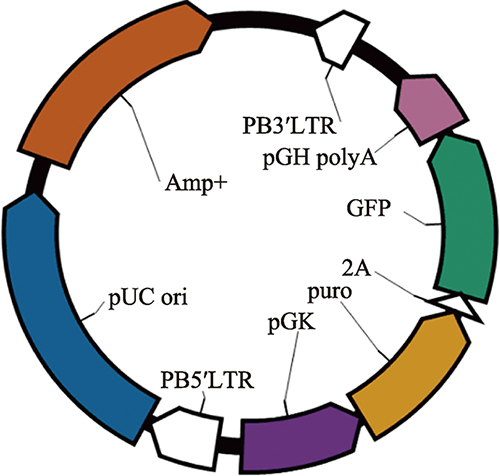
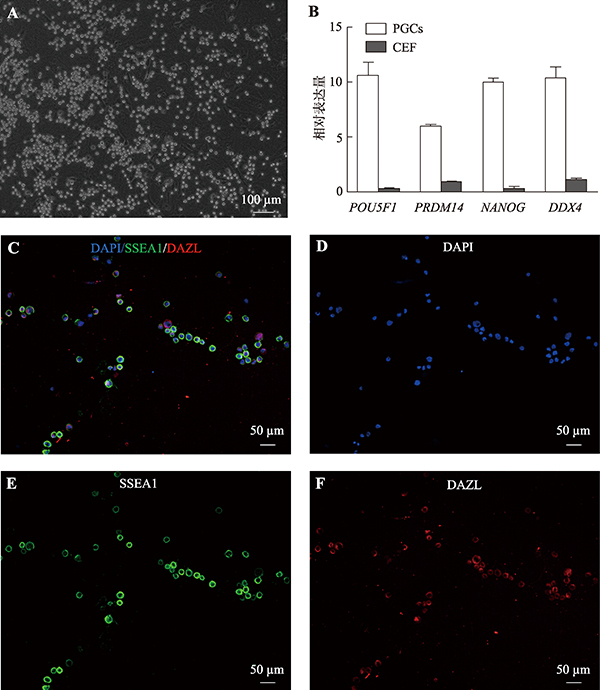
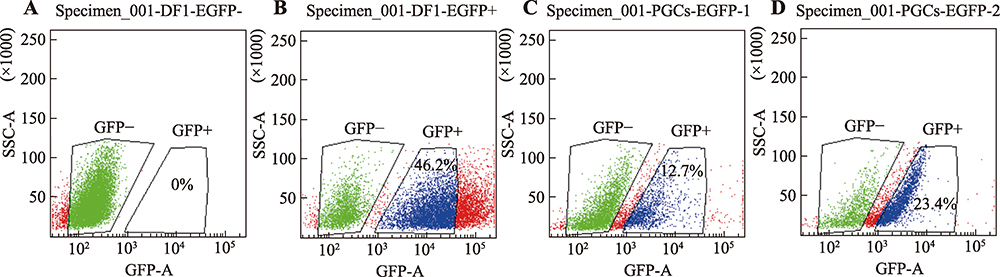
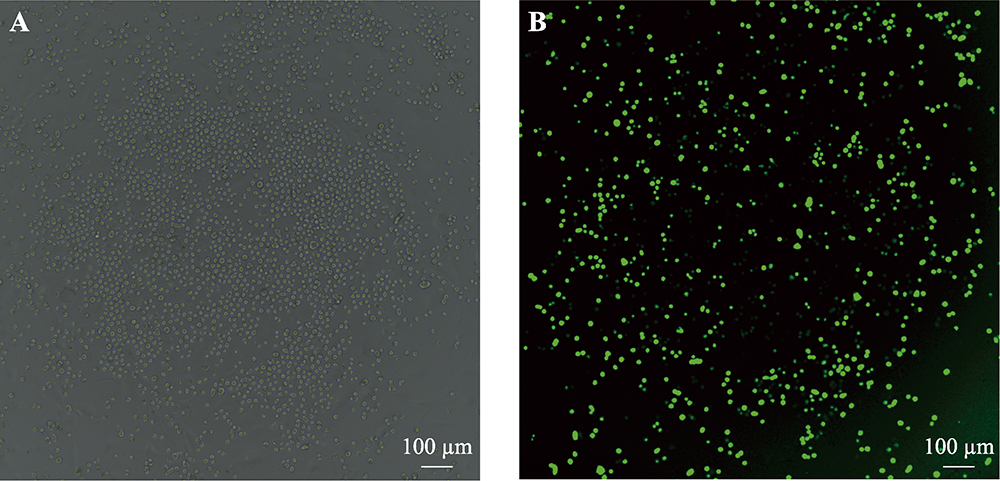
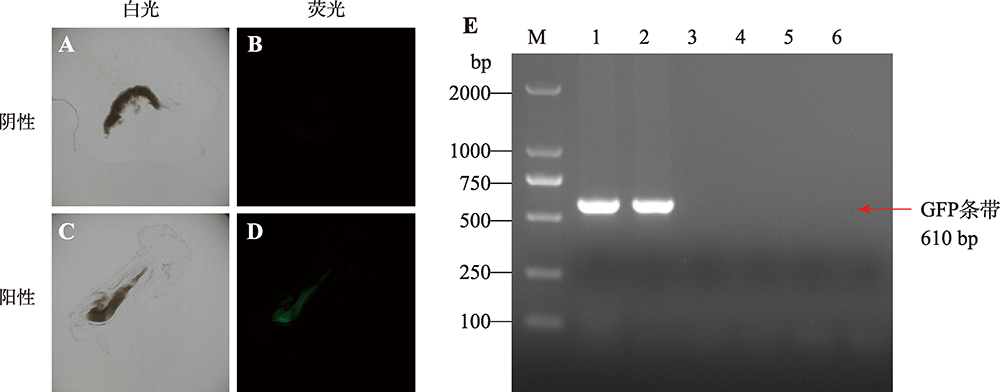
Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (3): 280-288.doi: 10.16288/j.yczz.20-212
Optimization of transfection conditions of chicken primordial germ cells
Xian Zou1( ), Yanhua He1, Jingyi He1, Yan Wang1, Dingming Shu1(
), Yanhua He1, Jingyi He1, Yan Wang1, Dingming Shu1( ), Chenglong Luo1(
), Chenglong Luo1( )
)
- 1 State Key Laboratory of Livestock and Poultry Breeding & Guangdong Key Laboratory of Animal Breeding and Nutrition, Institute of Animal Science, Guangdong Academy of Agricultural Sciences, Guangzhou 510640, China
-
Received:2020-07-07Online:2021-03-16Published:2021-01-29 -
Supported by:the National Natural Science Foundation of Guangdong Province(2018B030311044);the Science and Technology Program of Guangdong Province(2017B020232003);the Science and Technology Program of Guangdong Province(2019A050505007);2020 Provincial Modern Agricultural Science and Technology Alliance Construction of Common Key Technology Innovation Team Project(2020KJ106);the Earmarked Fund for Modern Agro-Industry Technology Research System(CARS-41);Special Fund for Scientific Innovation Strategy-construction of High Level Academy of Agriculture Science(R2017PY-QY007)
Cite this article
Xian Zou, Yanhua He, Jingyi He, Yan Wang, Dingming Shu, Chenglong Luo. Optimization of transfection conditions of chicken primordial germ cells[J]. Hereditas(Beijing), 2021, 43(3): 280-288.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Primer sequences for RT-qPCR detection used in this study"
| 基因 | 引物序列(5′→3′) | 扩增产物 长度(bp) |
|---|---|---|
| POU5F1 | CCACCCTCCCCACCTCTAC | 181 |
| TGTGCCAGCCTAACCTCCT | ||
| PRDM14 | TGTTCGCCTACCGCTACTACCG | 106 |
| AGTGCTGGCGGAGTGTGTGTG | ||
| NANOG | AGACCTCTCCTTGACCACA | 171 |
| CCACTCTCACCTTTATCCT | ||
| DDX4 | TCCCAGAGCCCACACAGATG | 124 |
| AAGTGATGCGCCCTCCTCTC | ||
| GAPDH | GGTGGTGCTAAGCGTGTTAT | 151 |
| ACCTCTGCCATCTCTCCACA |
Table 2
Number of positive cells after 48 hours transfection of PGCs with different transfection reagents and different doses of GFP plasmid (cell/well)"
| Lipofectamine 2000 | Lipofectamine 3000 | Lipofectamine LTX & Plus Reagent |
|---|---|---|
| A1 (20.30±1.70)b | B1 (50.40±7.10)c | C1 (30.00±5.50)c |
| A2 (40.50±3.50)a | B2 (102.10±3.10)b | C2 (55.90±4.40)b |
| A3 (46.50±5.10)a | B3 (278.50±10.90)a | C3 (80.90±8.10)a |
| A4 (40.70±10.80)a | B4 (75.20±8.60)b | C4 (63.30±9.20)b |
| [1] | Chojnacka-PuchtaL, SawickaD. CRISPR/Cas9 gene editing in a chicken model: current approaches and applications. J Appl Genet, 2020, 61(2): 221- 229. |
| [2] | ZhangG, LiC, LiQ, LiB, LarkinDM, LeeC, StorzJF, AntunesA, GreenwoldMJ, MeredithRW, ÖdeenA, CuiJ, ZhouQ, XuL, PanH, WangZ, JinL, ZhangP, HuH, YangW, HuJ, XiaoJ, YangZ, LiuY, XieQ, YuH, LianJ, WenP, ZhangF, LiH, ZengY, XiongZ, LiuS, ZhouL, HuangZ, AnN, WangJ, ZhengQ, XiongY, WangG, WangB, WangJ, FanY, DaFR, Alfaro-NúñezA, SchubertM, OrlandoL, MourierT, HowardJT, GanapathyG, PfenningA, WhitneyO, RivasMV, HaraE, SmithJ, FarreM, NarayanJ, SlavovG, RomanovMN, BorgesR, MachadoJP, KhanI, SpringerMS, GatesyJ, HoffmannFG, OpazoJC, HåstadO, SawyerRH, KimH, KimKW, KimHJ, ChoS, LiN, HuangY, BrufordMW, ZhanX, DixonA, BertelsenMF, DerryberryE, WarrenW, WilsonRK, LiS, RayDA, GreenRE, O'BrienSJ, GriffinD, JohnsonWE, HausslerD, RyderOA, WillerslevE, GravesGR, AlströmP, FjeldsåJ, MindellDP, EdwardsSV, BraunEL, RahbekC, BurtDW, HoudeP, ZhangY, YangH, WangJ, ConsortiumAG, JarvisED, GilbertMT, WangJ. Comparative genomics reveals insights into avian genome evolution and adaptation. Science, 2014, 346(6215):1311- 1320. |
| [3] | van de Lavoir MC, DiamondJH, LeightonPA, Mather-LoveC, HeyerBS, BradshawR, KerchnerA, HooiLT, GessaroTM, SwanbergSE, DelanyME, EtchesRJ. Germline transmission of genetically modified primordial germ cells. Nature, 2006, 441(7094):766- 769. |
| [4] | KoslováA, TrefilP, MucksováJ, ReinišováM, PlachýJ, KalinaJ, KučerováD, GerykJ, KrchlíkováV, LejčkováB, HejnarJ. Precise CRISPR/Cas9 editing of the NHE1 gene renders chickens resistant to the J subgroup of avian leukosis virus. Proc Natl Acad Sci USA, 2020, 117(4):2108- 2112. |
| [5] | LeeHJ, YoonJW, JungKM, KimYM, ParkJS, LeeKY, ParkKJ, HwangYS, ParkYH, RengarajD, HanJY. Targeted gene insertion into Z chromosome of chicken primordial germ cells for avian sexing model development. FASEB J, 2019, 33(7): 8519- 8529. |
| [6] | DimitrovL, PedersenD, ChingKH, YiH, CollariniEJ, IzquierdoS, vande Lavoir MC, LeightonPA. Germline gene editing in chickens by efficient CRISPR-mediated homologous recombination in primordial germ cells. PLoS One, 2016, 11(4): e154303. |
| [7] | OishiI, YoshiiK, MiyaharaD, KagamiH, TagamiT. Targeted mutagenesis in chicken using CRISPR/Cas9 system. Sci Rep, 2016, 6: 23980. |
| [8] | ChenMJ, ChenDY, XieL, LuZP, YangMM, MoLF, LUKH, LuYQ. Effect of recipient embryos age on homing of chicken primordial germ cell after transplantation. Journal of Southern Agriculture, 2016, 47(06): 1014- 1018. |
| 陈美娟, 陈东阳, 谢龙, 陆振萍, 杨蒙蒙, 莫丽芬, 卢克焕, 陆阳清. 受体胚龄对鸡原始生殖细胞移植后归巢的影响. 南方农业学报, 2016, 47(06): 1014- 1018. | |
| [9] | LillicoSG, McGrewMJ, ShermanA, SangHM. Transgenic chickens as bioreactors for protein-based drugs. Drug Discov Today, 2005, 10(3):191- 196. |
| [10] | IntarapatS, SternCD. Chick stem cells: Current progress and future prospects. Stem Cell Res, 2013, 11(3):1378- 1392. |
| [11] | LeeHJ, LeeHC, HanJY. Germline modification and engineering in avian species. Mol Cells, 2015, 38(9):743- 749. |
| [12] | MacdonaldJ, TaylorL, ShermanA, KawakamiK, TakahashiY, SangHM, McGrewMJ. Efficient genetic modification and germ-line transmission of primordial germ cells using piggyBac and Tol2 transposons. Proc Natl Acad Sci USA, 2012,109(23): E1466-E1472. |
| [13] | HeCW, HeYL, WuYH, ShaoFH. Effects of different transfection reagents on lentiviral packaging efficiency and the lentiviral infection efficiency of guinea-pig fibroblasts after packaging. Heilongjiang Anim Sci Veter Med, 2015, ( 10): 34- 38. |
| 何承文, 何玉龙, 吴月红, 邵风慧. 不同转染试剂对慢病毒包装及包装后病毒感染豚鼠成纤维细胞效率的影响. 黑龙江畜牧兽医, 2015, ( 10): 34- 38. | |
| [14] | ZhongCL, LiGL, MoJX, QuanR, WangHQ, LiZC, WuZF, ZhangXW. Effects of parameters, plasmid dosages and topological structures on transfection efficiency of porcine fetal fibroblasts using different electroporators. Hereditas (Beijing), 2017, 39(10): 930- 938. |
| 钟翠丽, 李国玲, 莫健新, 全绒, 王豪强, 李紫聪, 吴珍芳, 张献伟. 不同电转仪的电转参数、质粒用量和拓扑结构对猪胎儿成纤维细胞转染效率的影响. 遗传, 2017, 39(10): 930- 938. | |
| [15] | MacdonaldJ, GloverJD, TaylorL, SangHM, McGrewMJ. Characterisation and germline transmission of cultured avian primordial germ cells. PLoS One, 2010, 5(11): e15518. |
| [16] | ChoiJW, KimS, KimTM, KimYM, SeoHW, ParkTS, JeongJW, SongG, HanJY. Basic fibroblast growth factor activates MEK/ERK cell signaling pathway and stimulates the proliferation of chicken primordial germ cells. PLoS One, 2010, 5(9): e12968. |
| [17] | ParkTS, HanJY. piggyBac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proc Natl Acad Sci USA, 2012, 109(24):9337- 9341. |
| [18] | YamamotoY, UsuiF, NakamuraY, ItoY, TagamiT, NirasawaK, MatsubaraY, OnoT, KagamiH. A novel method to isolate primordial germ cells and its use for the generation of germline chimeras in chicken. Biol Reprod, 2007, 77(1): 115- 119. |
| [19] | WangL, ChenMJ, ChenDY, PengSF, ZhouXL, LiaoYY, YangXG, XuHY, LuSS, ZhangM, LuKH, LuYQ. Derivation and characterization of primordial germ cells from Guangxi yellow-feather chickens. Poult Sci, 2017, 96(5):1419- 1425. |
| [20] | TaylorL, CarlsonDF, NandiS, ShermanA, FahrenkrugSC, McGrewMJ. Efficient TALEN-mediated gene targeting of chicken primordial germ cells. Development, 2017, 144(5):928- 934. |
| [21] | NaitoM, HarumiT, KuwanaT. Long-term culture of chicken primordial germ cells isolated from embryonic blood and production of germline chimaeric chickens. Anim Reprod Sci, 2015, 153:50- 61. |
| [1] | Zhihui Gao, Jiaxin Huang, Haoyu Luo, Haidong Xu, Ming Lou, Bolin Ning, Xiaoxu Xing, Fang Mu, Hui Li, Ning Wang. Characterization of the genomic and transcriptional structure of chicken NRG4 gene [J]. Hereditas(Beijing), 2023, 45(5): 447-458. |
| [2] | Bingyuan Wang, Yulian Mu, Kui Li, Zhiguo Liu. Research progress of stem cells in agricultural animals [J]. Hereditas(Beijing), 2020, 42(11): 1073-1080. |
| [3] | Qiying Leng, Jiahui Zheng, Haidong Xu, Patricia Adu-Asiamah, Ying Zhang, Bingwang Du, Li Zhang. Cloning and expression analysis of chicken circular transcript of insulin degrading enzyme gene [J]. Hereditas(Beijing), 2019, 41(12): 1129-1137. |
| [4] | Jiahui Chen, Xueyi Ren, min Li, Shiyi Lu, Tian Cheng, Liangtian Tan, Shaodong Liang, Danlin He, Qingbin Luo, Qinghua Nie, Xiquan Zhang, Wen Luo. The cell cycle pathway regulates chicken abdominal fat deposition as revealed by transcriptome sequencing [J]. Hereditas(Beijing), 2019, 41(10): 962-973. |
| [5] | Yankai Chu,Yanfei Jin,Tianyu Xing,Guangwei Ma,Tingting Cui,Xiaohong Yan,Hui Li,Ning Wang. Post-transcriptional regulation of chicken PPARγ transcript variant 3 by upstream open reading frame [J]. Hereditas(Beijing), 2018, 40(8): 657-667. |
| [6] | Xiaofei Zhang,He Song,Jing Liu,Wenjian Zhang,Xiaohong Yan,Hui Li,Ning Wang. Identification and analysis of ZFPM2 as a target gene of miR-17-92 cluster in chicken [J]. Hereditas(Beijing), 2017, 39(4): 333-345. |
| [7] | Guangqi Li, Congjiao Sun, Guiqin Wu, Fengying Shi, Aiqiao Liu, Hao Sun, Ning Yang. Transcriptome sequencing identifies potential regulatory genes involved in chicken eggshell brownness [J]. Hereditas(Beijing), 2017, 39(11): 1102-1111. |
| [8] | Min Cheng, Wenjian Zhang, Tianyu Xing, Xiaohong Yan, Yumao Li, Hui Li, Ning Wang. Functional analysis of the upstream regulatory region of chicken miR-17-92 cluster [J]. Hereditas(Beijing), 2016, 38(8): 724-735. |
| [9] | Tao Zhang, Wenhao Wang, Genxi Zhang, Jinyu Wang, Qian Xue, Yuping Gu. A genome-wide association study on body weight traits of Jinghai yellow chicken [J]. HEREDITAS(Beijing), 2015, 37(8): 811-820. |
| [10] | Junping Ma, Xi Yang, Na Lv, Fei Liu, Yan Chen, Baoli Zhu. Re-sequencing and assembly of chicken T cell receptor gamma locus [J]. HEREDITAS(Beijing), 2015, 37(6): 568-574. |
| [11] | Xiaoyan Song, Dexiang Zhang, Wenwu Zhang, Congliang Ji, Xiquan Zhang, Qingbin Luo. Gene expression profiling of three tissues of chicken after heat stress treatment by microarray technique [J]. HEREDITAS(Beijing), 2014, 36(8): 800-808. |
| [12] | Jinlong Li, Shaoqing Tang, Zhiyuan Zou, Haichao Wang, Qing Xu. Establishment of fluorescence labeling and capillary electrophoresis in MSAP for Beijing You chicken [J]. HEREDITAS(Beijing), 2014, 36(5): 495-502. |
| [13] | MU Ren BIAN Yan-Chao PU Ya-Bin LI Xiang-Chen WANG Feng-Long GUAN Wei-Jun. Isolation and biological characterization of mesenchymal stem cells from Beijing fatty chicken fetal liver [J]. HEREDITAS, 2013, 35(3): 365-372. |
| [14] | WANG Li, NA Wei, WANG Yu-Xiang, WANG Yan-Bo, WANG Ning, WANG Qi-Gui, LI Yu-Mao, LI Hui. Characterization of chicken PPARγ expression and its impact on adipocyte proliferation and differentiation [J]. HEREDITAS, 2012, 34(4): 454-464. |
| [15] | . Association of single nucleotide polymorphism of RB1 gene with body weight traits in chicken [J]. HEREDITAS, 2012, 34(10): 1320-1327. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||