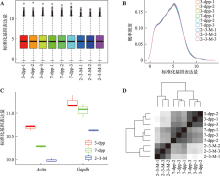
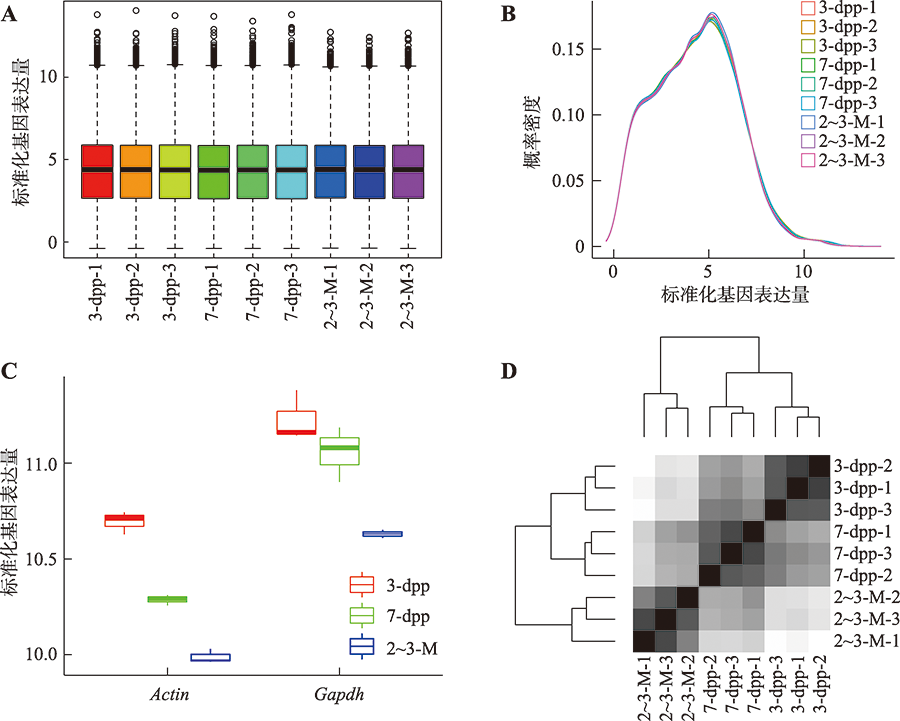
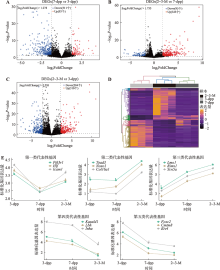
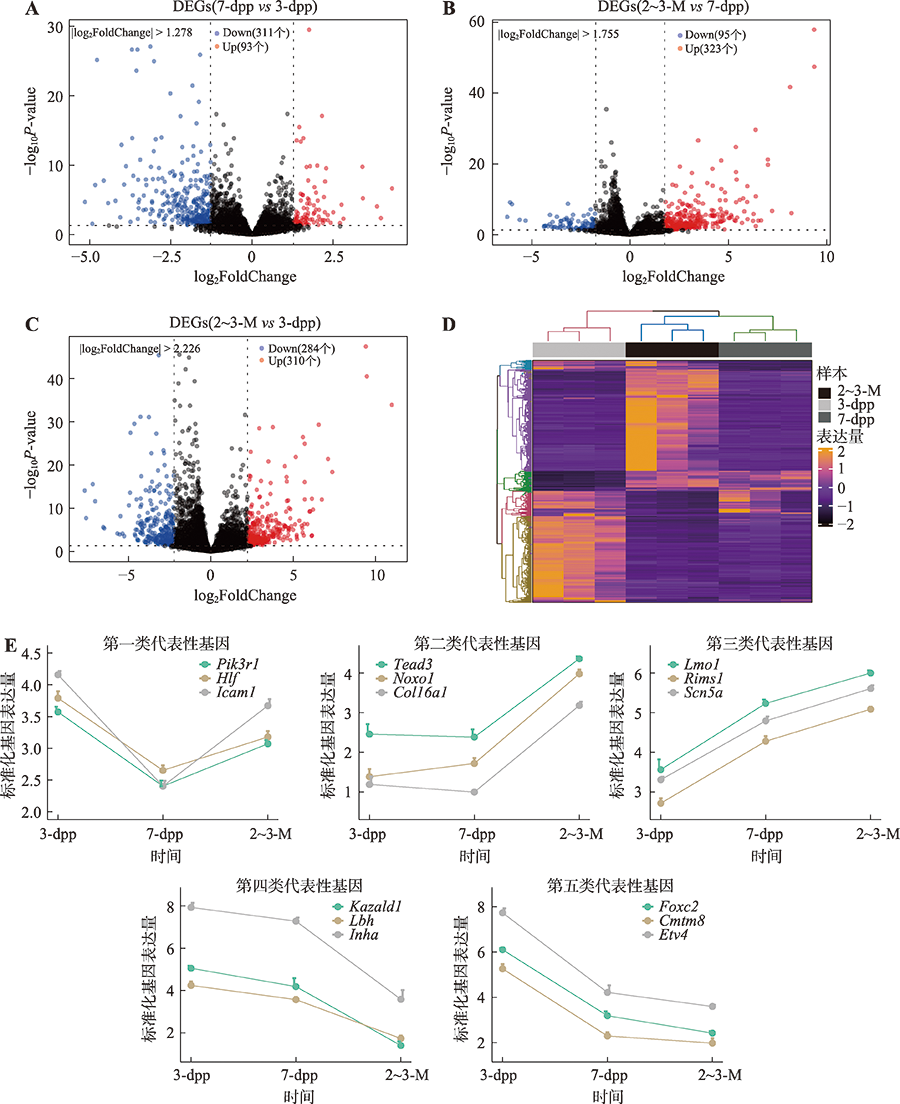
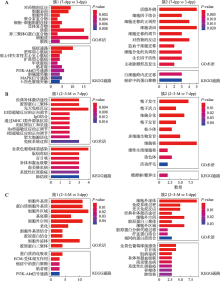
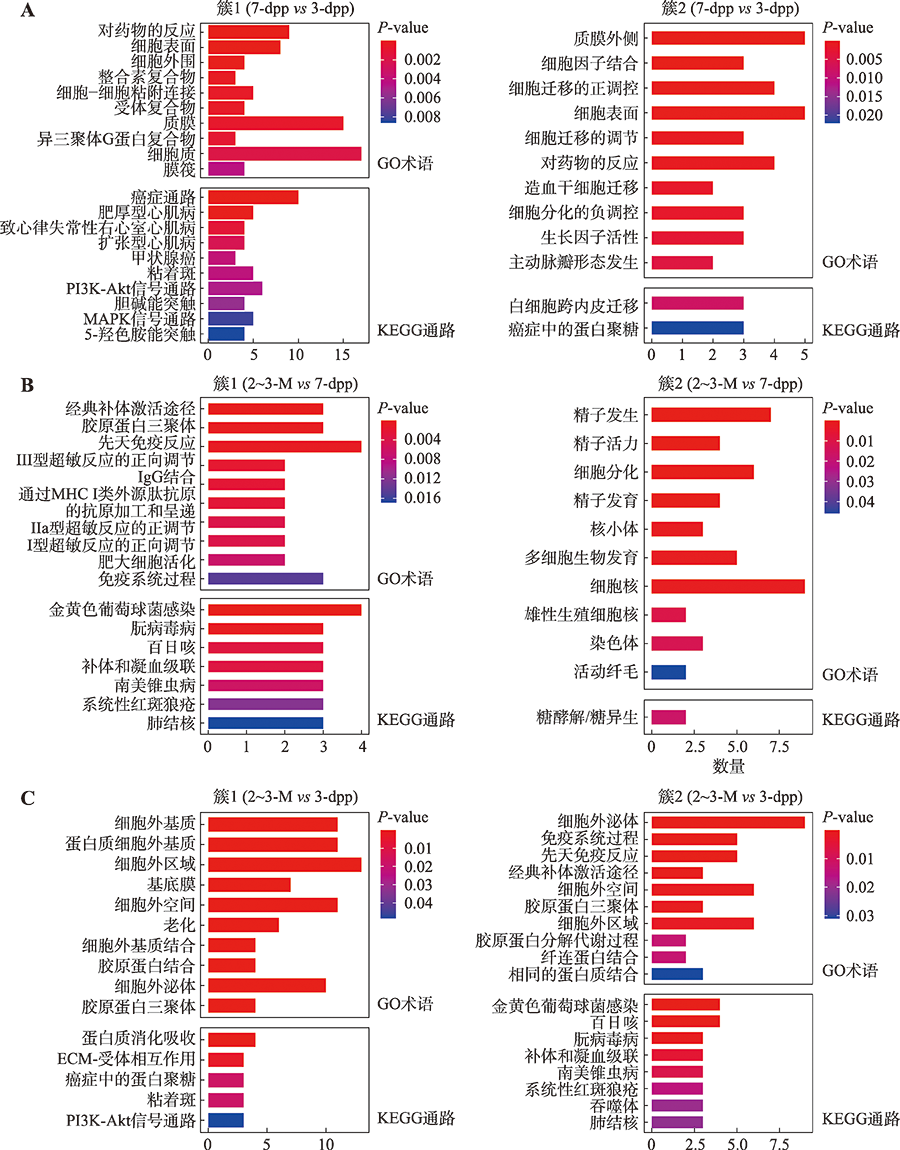
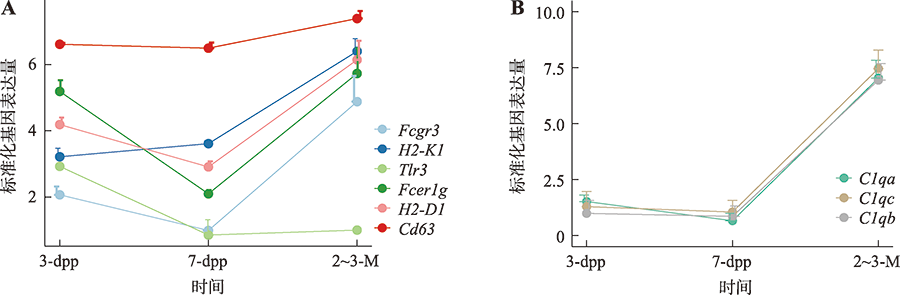
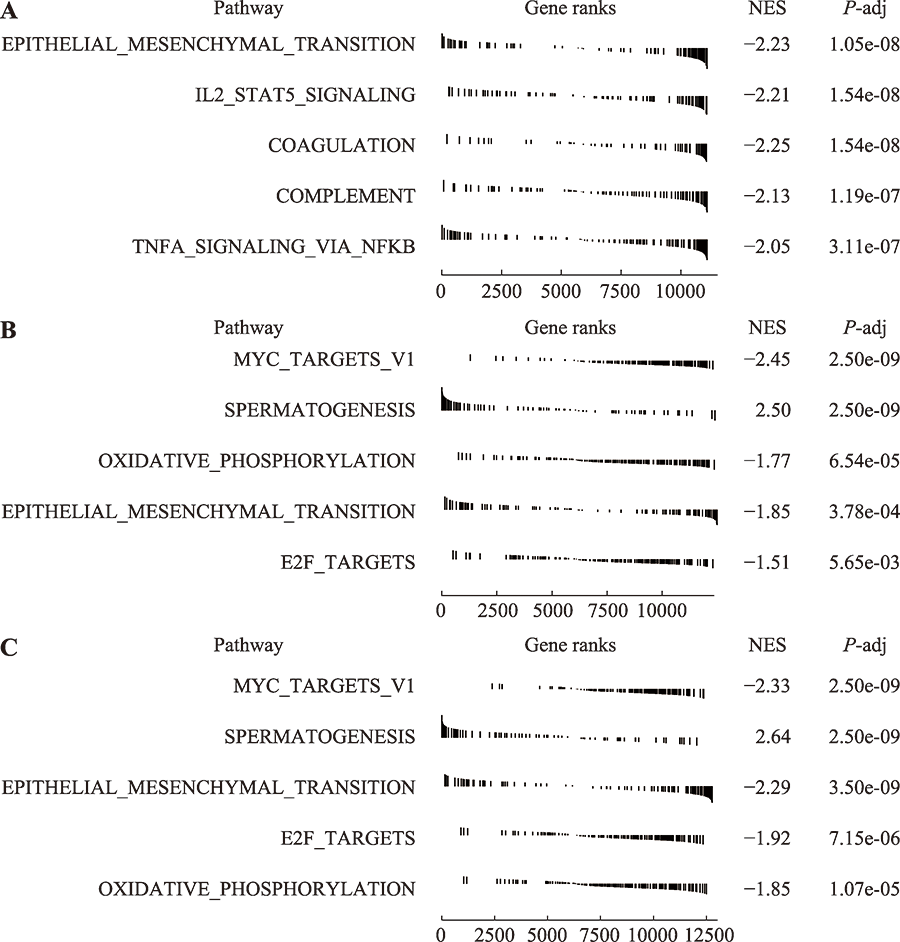
Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (7): 591-608.doi: 10.16288/j.yczz.22-047
• Research Article • Previous Articles Next Articles
Transcriptome analysis of mouse male germline stem cells reveals characteristics of mature spermatogonial stem cells
Yan Guo1( ), Lele Yang2(
), Lele Yang2( ), Huayu Qi2(
), Huayu Qi2( )
)
- 1. Department of Pharmaceutical Biotechnology, School of Life Sciences, Department of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230026, China
2. Center for Cell Lineage and Development,Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, Guangzhou 510530, China
-
Received:2022-02-23Revised:2022-04-26Online:2022-07-20Published:2022-07-06 -
Contact:Yang Lele,Qi Huayu E-mail:rxm749426464@qq.com;yang_lele@gibh.ac.cn;qi_huayu@gibh.ac.cn -
Supported by:Supported by the Natural Science Foundation of Guangdong Province No(2020A1515010882);Science and Technology Planning Project of Guangdong Province No(2020B1212060052);the Frontier Research Program of Guangzhou Regenerative Medicine and Health, Bioland Laboratory No(2018GZR110105021)
Cite this article
Yan Guo, Lele Yang, Huayu Qi. Transcriptome analysis of mouse male germline stem cells reveals characteristics of mature spermatogonial stem cells[J]. Hereditas(Beijing), 2022, 44(7): 591-608.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
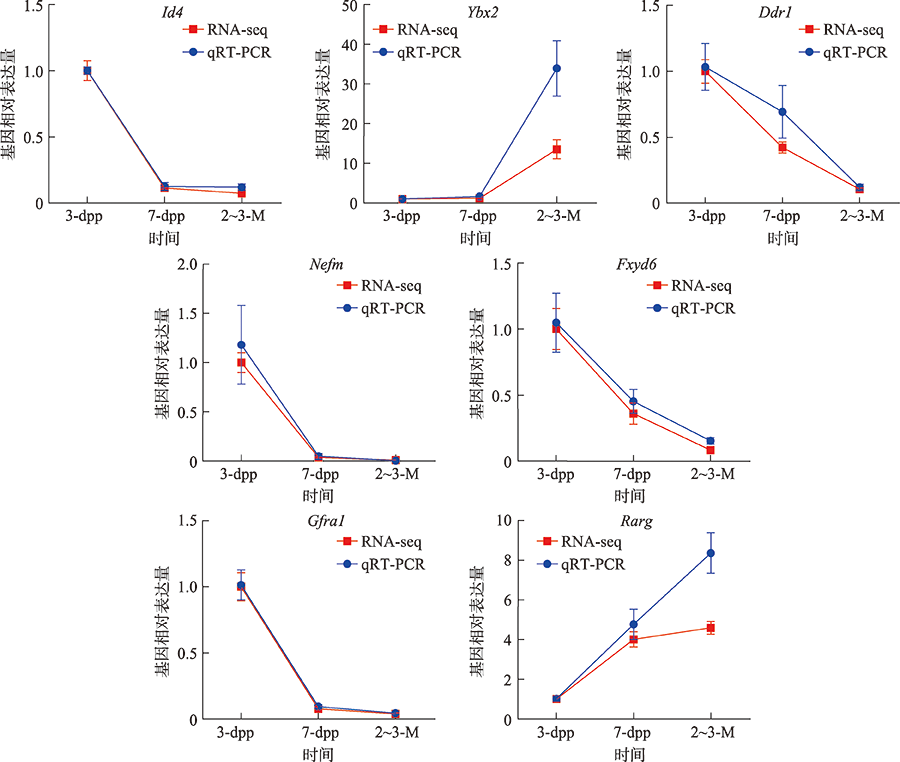
Supplementary Table 1
Primer sequences for qRT-PCR"
| 序号 | 基因 | 上游引物(5′→3′) | 下游引物(5′→3′) |
|---|---|---|---|
| 1 | Actin | TGTACCCAGGCATTGCTGACAG | CTGCTGGAAGGTGGACAGTG |
| 2 | Id4 | CACTCACCGCGCTCAACA | CTCCGGTGGCTTGTTTCTCTT |
| 3 | Ybx2 | GGAGTTTGATGTCGTGGAAGG | CGTCGATTAGGGGCATAGCG |
| 4 | Ddr1 | ATGCTGACATGAAGGGACATTT | GGTGTAGCCTACGAAGGTCCA |
| 5 | Nefm | ATGACGAGCCATTTCCCACT | TGCAGTCCAAGAGCATCGAG |
| 6 | Fxyd6 | TCTCCGTTGGGATACTTCTCAT | CTCCGCAGCGTTTGTAGTGAT |
| 7 | Gfra1 | AACTGCCAGCCAGAGTCAAG | GGCTGCTGGAGTCTATGTAG |
| 8 | Rarg | TGCCTGGTTTTACAGGGCTC | TCCGAGAATGTCATAGTGTCCT |
| [1] |
Vander Borght M, Wyns C. Fertility and infertility: definition and epidemiology. Clin Biochem, 2018, 62:2-10.
doi: 10.1016/j.clinbiochem.2018.03.012 |
| [2] |
Agarwal A, Baskaran S, Parekh N, Cho CL, Henkel R, Vij S, Arafa M, Panner Selvam MK, Shah R. Male infertility. Lancet, 2021, 397(10271):319-333.
doi: 10.1016/S0140-6736(20)32667-2 pmid: 33308486 |
| [3] |
Phillips BT, Gassei K, Orwig KE. Spermatogonial stem cell regulation and spermatogenesis. Philos Trans R Soc Lond B Biol Sci, 2010, 365(1546):1663-1678.
doi: 10.1098/rstb.2010.0026 |
| [4] | Mei XX, Wang J, Wu J. Extrinsic and intrinsic factors controlling spermatogonial stem cell self-renewal and differentiation. Asian J Androl, 2015, 17(3):347-354. |
| [5] | Jan SZ, Vormer TL, Jongejan A, Röling MD, Silber SJ, de Rooij DG, Hamer G, Repping S, van Pelt AMM. Unraveling transcriptome dynamics in human spermatogenesis. Development, 2017, 144(20):3659-3673. |
| [6] |
Izadyar F, Den Ouden K, Stout TAE, Stout J, Coret J, Lankveld DPK, Spoormakers TJP, Colenbrander B, Oldenbroek JK, Van der Ploeg KD, Woelders H, Kal HB, De Rooij DG. Autologous and homologous transplantation of bovine spermatogonial stem cells. Reproduction, 2003, 126(6):765-774.
pmid: 14748695 |
| [7] |
Schlatt S, Foppiani L, Rolf C, Weinbauer GF, Nieschlag E. Germ cell transplantation into X-irradiated monkey testes. Hum Reprod, 2002, 17(1):55-62.
pmid: 11756362 |
| [8] |
Jahnukainen K, Ehmcke J, Quader MA, Saiful Huq M, Epperly MW, Hergenrother S, Nurmio M, Schlatt S. Testicular recovery after irradiation differs in prepubertal and pubertal non-human primates, and can be enhanced by autologous germ cell transplantation. Hum Reprod, 2011, 26(8):1945-1954.
doi: 10.1093/humrep/der160 |
| [9] |
Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, Valli H, Rodriguez M, Ezzelarab M, Dargo G, Peterson K, Masterson K, Ramsey C, Ward T, Lienesch M, Volk A, Cooper DK, Thomson AW, Kiss JE, Penedo MCT, Schatten GP, Mitalipov S, Orwig KE. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell, 2012, 11(5):715-726.
doi: 10.1016/j.stem.2012.07.017 pmid: 23122294 |
| [10] |
Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science, 2010, 328(5974):62-67.
doi: 10.1126/science.1182868 |
| [11] |
Carrieri C, Comazzetto S, Grover A, Morgan M, Buness A, Nerlov C, O'Carroll D. A transit-amplifying population underpins the efficient regenerative capacity of the testis. J Exp Med, 2017, 214(6):1631-1641.
doi: 10.1084/jem.20161371 |
| [12] |
La HM, Mäkelä JA, Chan AL, Rossello FJ, Nefzger CM, Legrand JMD, De Seram M, Polo JM, Hobbs RM. Identification of dynamic undifferentiated cell states within the male germline. Nat Commun, 2018, 9(1):2819.
doi: 10.1038/s41467-018-04827-z |
| [13] |
Clevers H, Watt FM. Defining adult stem cells by function, not by phenotype. Annu Rev Biochem, 2018, 87:1015-1027.
doi: 10.1146/annurev-biochem-062917-012341 pmid: 29494240 |
| [14] |
Mäkelä JA, Hobbs RM. Molecular regulation of spermatogonial stem cell renewal and differentiation. Reproduction, 2019, 158(5):R169-R187.
doi: 10.1530/REP-18-0476 |
| [15] |
Nagano M, Avarbock MR, Brinster RL. Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol Reprod, 1999, 60(6):1429-1436.
pmid: 10330102 |
| [16] |
Nagano MC. Homing efficiency and proliferation kinetics of male germ line stem cells following transplantation in mice. Biol Reprod, 2003, 69(2):701-707.
doi: 10.1095/biolreprod.103.016352 |
| [17] |
Forbes CM, Flannigan R, Schlegel PN. Spermatogonial stem cell transplantation and male infertility: current status and future directions. Arab J Urol, 2017, 16(1):171-180.
doi: 10.1016/j.aju.2017.11.015 |
| [18] |
Schmidt JA, Abramowitz LK, Kubota H, Wu X, Niu Z, Avarbock MR, Tobias JW, Bartolomei MS, Brinster RL. In vivo and in vitro aging is detrimental to mouse spermatogonial stem cell function. Biol Reprod, 2011, 84(4):698-706.
doi: 10.1095/biolreprod.110.088229 |
| [19] |
Kanatsu-Shinohara M, Ogonuki N, Iwano T, Lee J, Kazuki Y, Inoue K, Miki H, Takehashi M, Toyokuni S, Shinkai Y, Oshimura M, Ishino F, Ogura A, Shinohara T. Genetic and epigenetic properties of mouse male germline stem cells during long-term culture. Development, 2005, 132(18):4155-4163.
pmid: 16107472 |
| [20] |
Subash SK, Kumar PG. Spermatogonial stem cells: a story of self-renewal and differentiation. Front Biosci (Landmark Ed), 2021, 26:163-205.
doi: 10.2741/4891 pmid: 33049667 |
| [21] |
Zhao X, Yang HQ. Progress on spermatogonial stem cells of large animals. Hereditas(Beijing), 2019, 41(8):686-702.
doi: 10.16288/j.yczz.19-167 pmid: 31447420 |
|
赵鑫, 杨化强. 大动物精原干细胞研究进展. 遗传, 2019, 41(8):686-702.
doi: 10.16288/j.yczz.19-167 pmid: 31447420 |
|
| [22] | Mäkelä JA, Toppari J. Spermatogenesis. In: Simoni M, Huhtaniemi I, eds. Endocrinology of the Testis and Male Reproduction. Endocrinology. Springer Cham, 2017, 417-455. |
| [23] |
Liao JY, Suen HC, Luk ACS, Yang LL, Lee AWT, Qi HY, Lee TL. Transcriptomic and epigenomic profiling of young and aged spermatogonial stem cells reveals molecular targets regulating differentiation. PLoS Genet, 2021, 17(7):e1009369.
doi: 10.1371/journal.pgen.1009369 |
| [24] |
Yang LL, Wu W, Qi HY. Gene expression profiling revealed specific spermatogonial stem cell genes in mouse. Genesis, 2013, 51(2):83-96.
doi: 10.1002/dvg.22358 |
| [25] |
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods, 2015, 12(4):357-360.
doi: 10.1038/NMETH.3317 |
| [26] |
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 2014, 15(12):550.
doi: 10.1186/s13059-014-0550-8 |
| [27] |
Jiang YA, Leng JX, Lin QX, Zhou F. Epithelial- mesenchymal transition related genes in unruptured aneurysms identified through weighted gene coexpression network analysis. Sci Rep, 2022, 12(1):225.
doi: 10.1038/s41598-021-04390-6 |
| [28] |
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res, 2021, 49(D1):D605-D612.
doi: 10.1093/nar/gkaa1074 pmid: 33237311 |
| [29] |
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res, 2003, 13(11):2498-2504.
doi: 10.1101/gr.1239303 pmid: 14597658 |
| [30] |
Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics, 2003, 4:2.
pmid: 12525261 |
| [31] |
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol, 2003, 4(5):P3.
pmid: 12734009 |
| [32] |
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics, 2011, 27(12):1739-1740.
doi: 10.1093/bioinformatics/btr260 pmid: 21546393 |
| [33] |
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA, 2005, 102(43):15545-15550.
doi: 10.1073/pnas.0506580102 |
| [34] |
Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst, 2015, 1(6):417-425.
doi: 10.1016/j.cels.2015.12.004 pmid: 26771021 |
| [35] |
Liberzon A. A description of the Molecular Signatures Database (MSigDB) Web site. Methods Mol Biol, 2014, 1150:153-160.
doi: 10.1007/978-1-4939-0512-6_9 pmid: 24743996 |
| [36] | Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, 1995, 57(1):289-300. |
| [37] |
Wang T, Zhang XY, Chen QY, Deng TT, Zhang Y, Li N, Shang T, Chen YM, Han DS. Toll-like receptor 3-initiated antiviral responses in mouse male germ cells in vitro. Biol Reprod, 2012, 86(4):106.
doi: 10.1095/biolreprod.111.096719 pmid: 22262694 |
| [38] | Chen QY, Zhu WW, Liu ZH, Yan KQ, Zhao ST, Han DS. Toll-like receptor 11-initiated innate immune response in male mouse germ cells. Biol Reprod, 2014, 90(2):38. |
| [39] |
Mathieu M, Névo N, Jouve M, Valenzuela JI, Maurin M, Verweij FJ, Palmulli R, Lankar D, Dingli F, Loew D, Rubinstein E, Boncompain G, Perez F, Théry C. Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nat Commun, 2021, 12(1):4389.
doi: 10.1038/s41467-021-24384-2 |
| [40] |
Mangold CA, Masser DR, Stanford DR, Bixler GV, Pisupati A, Giles CB, Wren JD, Ford MM, Sonntag WE, Freeman WM. CNS-wide sexually dimorphic induction of the major histocompatibility Complex 1 pathway with aging. J Gerontol A Biol Sci Med Sci, 2017, 72(1):16-29.
doi: 10.1093/gerona/glv232 |
| [41] |
Son M, Diamond B, Santiago-Schwarz F. Fundamental role of C1q in autoimmunity and inflammation. Immunol Res, 2015, 63(1-3):101-106.
doi: 10.1007/s12026-015-8705-6 |
| [42] |
Kanatsu-Shinohara M, Ogonuki N, Matoba S, Ogura A, Shinohara T. Autologous transplantation of spermatogonial stem cells restores fertility in congenitally infertile mice. Proc Natl Acad Sci USA, 2020, 117(14):7837-7844.
doi: 10.1073/pnas.1914963117 |
| [43] |
Lord T, Nixon B. Metabolic changes accompanying spermatogonial stem cell differentiation. Dev Cell, 2020, 52(4):399-411.
doi: 10.1016/j.devcel.2020.01.014 |
| [44] |
Park MH, Park JE, Kim MS, Lee KY, Hwang JY, Yun JI, Choi JH, Lee E, Lee ST. Effects of extracellular matrix protein-derived signaling on the maintenance of the undifferentiated state of spermatogonial stem cells from porcine neonatal testis. Asian-Australas J Anim Sci, 2016, 29(10):1398-1406.
doi: 10.5713/ajas.15.0856 |
| [45] |
Shinohara T, Avarbock MR, Brinster RL. Beta1- and alpha6-integrin are surface markers on mouse spermatogonial stem cells. Proc Natl Acad Sci USA, 1999, 96(10):5504-5509.
doi: 10.1073/pnas.96.10.5504 |
| [46] |
Domke LM, Rickelt S, Dörflinger Y, Kuhn C, Winter-Simanowski S, Zimbelmann R, Rosin-Arbesfeld R, Heid H, Franke WW. The cell-cell junctions of mammalian testes: I. The adhering junctions of the seminiferous epithelium represent special differentiation structures. Cell Tissue Res, 2014, 357(3):645-665.
doi: 10.1007/s00441-014-1906-9 pmid: 24907851 |
| [47] |
Morimoto H, Ogonuki N, Kanatsu-Shinohara M, Matoba S, Ogura A, Shinohara T. Spermatogonial stem cell transplantation into nonablated mouse recipient testes. Stem Cell Reports, 2021, 16(7):1832-1844.
doi: 10.1016/j.stemcr.2021.05.013 pmid: 34143973 |
| [48] |
Zhao ST, Zhu WW, Xue SP, Han DS. Testicular defense systems: immune privilege and innate immunity. Cell Mol Immunol, 2014, 11(5):428-437.
doi: 10.1038/cmi.2014.38 |
| [49] |
Yule TD, Montoya GD, Russell LD, Williams TM, Tung KS. Autoantigenic germ cells exist outside the blood testis barrier. J Immunol, 1988, 141(4):1161-1167.
pmid: 3397538 |
| [50] |
Setchell BP. The testis and tissue transplantation: historical aspects. J Reprod Immunol, 1990, 18(1):1-8.
pmid: 2213727 |
| [51] |
Formosa LE, Ryan MT. Mitochondrial OXPHOS complex assembly lines. Nat Cell Biol, 2018, 20(5):511-513.
doi: 10.1038/s41556-018-0098-z pmid: 29662174 |
| [52] |
Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen-Orr S, Laevsky I, Amit M, Bomze D, Elena-Herrmann B, Scherf T, Nissim-Rafinia M, Kempa S, Itskovitz-Eldor J, Meshorer E, Aberdam D, Nahmias Y. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab, 2015, 21(3):392-402.
doi: 10.1016/j.cmet.2015.02.002 pmid: 25738455 |
| [53] |
Voigt AL, Thiageswaran S, de Lima E Martins Lara N, Dobrinski I. Metabolic requirements for spermatogonial stem cell establishment and maintenance in vivo and in vitro. Int J Mol Sci, 2021, 22(4):1998.
doi: 10.3390/ijms22041998 |
| [54] | Brinster RL, Troike DE. Requirements for blastocyst development in vitro. J Anim Sci, 1979, 49Suppl 2: 26-34. |
| [55] |
Butcher L, Coates A, Martin KL, Rutherford AJ, Leese HJ. Metabolism of pyruvate by the early human embryo. Biol Reprod, 1998, 58(4):1054-1056.
pmid: 9546739 |
| [56] |
Gardner DK, Lane M, Stevens J, Schoolcraft WB. Noninvasive assessment of human embryo nutrient consumption as a measure of developmental potential. Fertil Steril, 2001, 76(6):1175-1180.
pmid: 11730746 |
| [57] |
Leese HJ, Barton AM. Pyruvate and glucose uptake by mouse ova and preimplantation embryos. J Reprod Fertil, 1984, 72(1):9-13.
pmid: 6540809 |
| [58] |
Leese HJ. Metabolism of the preimplantation embryo: 40 years on. Reproduction, 2012, 143(4):417-427.
doi: 10.1530/REP-11-0484 |
| [59] | Tischler J, Gruhn WH, Reid J, Allgeyer E, Buettner F, Marr C, Theis F, Simons BD, Wernisch L, Surani MA. Metabolic regulation of pluripotency and germ cell fate through α-ketoglutarate. EMBO J, 2019, 38(1):e99518. |
| [60] |
Yoshida S. Open niche regulation of mouse spermatogenic stem cells. Dev Growth Differ, 2018, 60(9):542-552.
doi: 10.1111/dgd.12574 |
| [61] |
Hayashi Y, Otsuka K, Ebina M, Igarashi K, Takehara A, Matsumoto M, Kanai A, Igarashi K, Soga T, Matsui Y. Distinct requirements for energy metabolism in mouse primordial germ cells and their reprogramming to embryonic germ cells. Proc Natl Acad Sci USA, 2017, 114(31):8289-8294.
doi: 10.1073/pnas.1620915114 |
| [62] | Voigt AL, Kondro DA, Powell D, Valli-Pulaski H, Ungrin M, Stukenborg JB, Klein C, Lewis IA, Orwig KE, Dobrinski I. Unique metabolic phenotype and its transition during maturation of juvenile male germ cells. FASEB J, 2021, 35(5):e21513. |
| [63] |
Sohni A, Tan K, Song HW, Burow D, de Rooij DG, Laurent L, Hsieh TC, Rabah R, Hammoud SS, Vicini E, Wilkinson MF. The neonatal and adult human testis defined at the single-cell level. Cell Rep, 2019, 26(6): 1501-1517.e4.
doi: 10.1016/j.celrep.2019.01.045 |
| [1] | Hongbo Luo, Pengbo Cao, Gangqiao Zhou. Prognostic and predictive value of a DNA methylation-driven transcriptional signature in hepatocellular carcinoma [J]. Hereditas(Beijing), 2020, 42(8): 775-787. |
| [2] | Tianpei Shi,Li Zhang. Application of whole transcriptomics in animal husbandry [J]. Hereditas(Beijing), 2019, 41(3): 193-205. |
| [3] | Gaohua Zhang, Shutao Yu, He Wang, Xuda Wang. Transcriptome profiling of high oleic peanut under low temperatureduring germination [J]. Hereditas(Beijing), 2019, 41(11): 1050-1059. |
| [4] | Jiahui Chen, Xueyi Ren, min Li, Shiyi Lu, Tian Cheng, Liangtian Tan, Shaodong Liang, Danlin He, Qingbin Luo, Qinghua Nie, Xiquan Zhang, Wen Luo. The cell cycle pathway regulates chicken abdominal fat deposition as revealed by transcriptome sequencing [J]. Hereditas(Beijing), 2019, 41(10): 962-973. |
| [5] | Lan Ren,Rudan Xiao,Qian Zhang,Xiaomin Lou,Zhaojun Zhang,Xiangdong Fang. Synergistic regulation of the erythroid differentiation of K562 cells by KLF1 and KLF9 [J]. Hereditas(Beijing), 2018, 40(11): 998-1006. |
| [6] | Yajun Liu,Feng Zhang,Hongde Liu,Xiao Sun. The application of next-generation sequencing techniques in studying transcriptional regulation in embryonic stem cells [J]. Hereditas(Beijing), 2017, 39(8): 717-725. |
| [7] | Kai Wei,Lei Ma. Concept development of housekeeping genes in the high-throughput sequencing era [J]. Hereditas(Beijing), 2017, 39(2): 127-134. |
| [8] | Guangqi Li, Congjiao Sun, Guiqin Wu, Fengying Shi, Aiqiao Liu, Hao Sun, Ning Yang. Transcriptome sequencing identifies potential regulatory genes involved in chicken eggshell brownness [J]. Hereditas(Beijing), 2017, 39(11): 1102-1111. |
| [9] | Yongming Liu, Ling Zhang, Tao Qiu, Zhuofan Zhao, Moju Cao. Research progress on mechanisms of male sterility in plants based on high-throughput RNA sequencing [J]. Hereditas(Beijing), 2016, 38(8): 677-687. |
| [10] | Xiao Zhang, Guifang Jia. RNA epigenetic modification: N6-methyladenosine [J]. HEREDITAS(Beijing), 2016, 38(4): 275-288. |
| [11] | Shuaiqi Zhu, Yifu Gong, Yuqing Hang, Hao Liu, Heyu Wang. Transcriptome analysis of Dunaliella viridis [J]. HEREDITAS(Beijing), 2015, 37(8): 828-836. |
| [12] | Dong Wang, Yongjun Li, Nan Ding, Junyun Wang, Qiong Yang, Yaran Yang, Yanming Li, Xiangdong Fang, Hua Zhao. Molecular networks and mechanisms of epithelial-mesenchymal transition regulated by miRNAs in the malignant melanoma cell line [J]. HEREDITAS(Beijing), 2015, 37(7): 673-682. |
| [13] | Wei Jiang, Yixing Fan, Xian Qiao, Yanjun Zhang, Zhihong Liu, Yanhong Zhao, Ruijun Wang, Zhixin Wang, Wenguang Zhang, Rui Su, Jinquan Li. The transcriptome research progresses of skin hair follicle development [J]. HEREDITAS(Beijing), 2015, 37(6): 528-534. |
| [14] | Yongming Liu, Ling Zhang, Jianyu Zhou, Moju Cao. Research progress of the bHLH transcription factors involved in genic male sterility in plants [J]. HEREDITAS(Beijing), 2015, 37(12): 1194-1203. |
| [15] | Xin Liu,Xueying Song,Xiaoping Zhang,Yinglun Han,Ting Zhu,Rong Xiao,Qingwei Li. Genetic basis of immune response of lymphocyte-like cells in the mucosal immune system of Lampetra japonica [J]. HEREDITAS(Beijing), 2015, 37(11): 1149-1159. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||