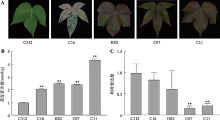
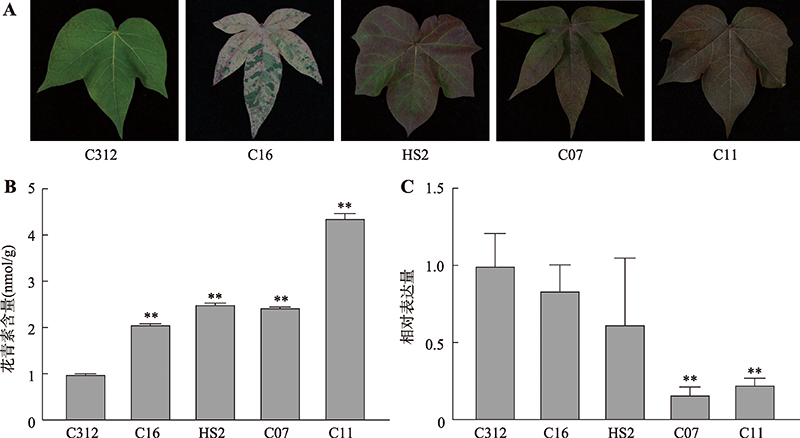
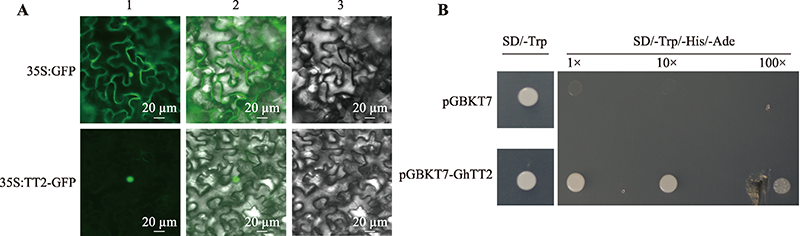
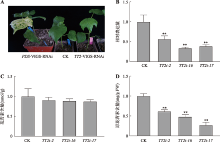
Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (8): 720-728.doi: 10.16288/j.yczz.22-100
• Research Article • Previous Articles
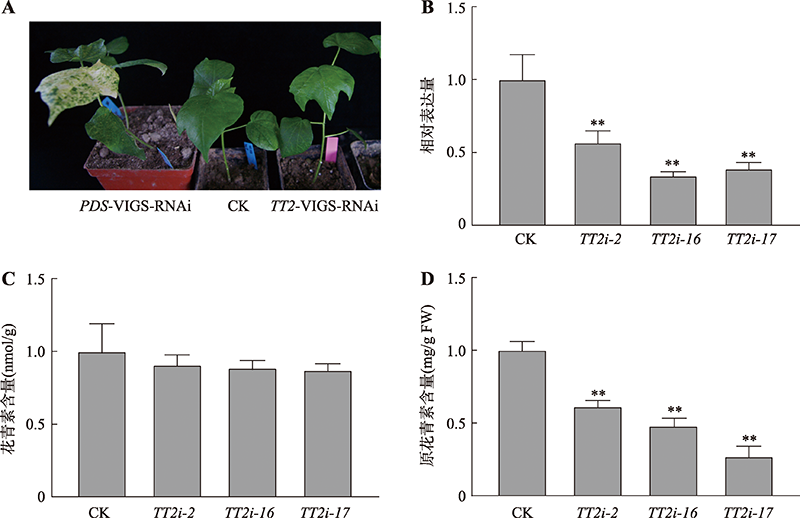
Cloning and characterization of the MYB transcription factor gene GhTT2 in Gossypium hirsutum
Rongrong Mu( ), Qingqing Niu, Yuqiang Sun, Jun Mei(
), Qingqing Niu, Yuqiang Sun, Jun Mei( ), Meng Miao(
), Meng Miao( )
)
- Plant Genomics and Molecular Improvement of Colored Fiber Lab, School of Life Sciences and Medicine, Zhejiang Sci-Tech University, Hangzhou 310018, China
-
Received:2022-04-06Revised:2022-05-24Online:2022-08-20Published:2022-06-29 -
Contact:Mei Jun,Miao Meng E-mail:rongrongmu1020xx@163.com;mj2019@zstu.edu.cn;miaom@whu.edu.cn -
Supported by:the National Natural Science Foundation of China(32001591);the Science Foundation of Zhejiang Sci-Tech University(19042401-Y)
Cite this article
Rongrong Mu, Qingqing Niu, Yuqiang Sun, Jun Mei, Meng Miao. Cloning and characterization of the MYB transcription factor gene GhTT2 in Gossypium hirsutum[J]. Hereditas(Beijing), 2022, 44(8): 720-728.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
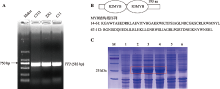
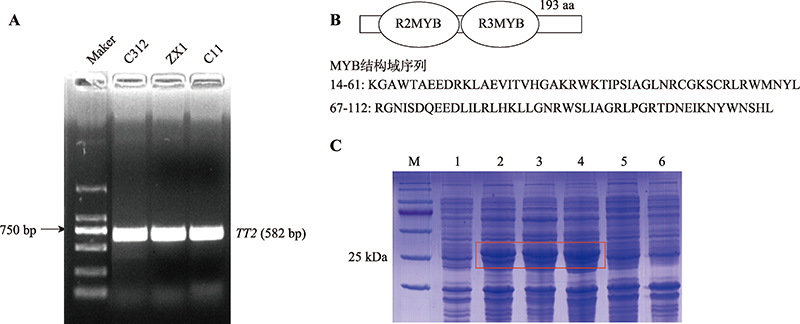
Table 1
Primers used in this study"
| 引物名称 | 引物序列(5′→3′) | 用途 |
|---|---|---|
| GhTT2 | F: CACCATGGAGCAAAATTTGTCCCT | 用于基因克隆、亚细胞定位及酵母转录激活载体构建 |
| R: AAACTTAAAACCGTCGTCAA | ||
| p28a-GhTT2 | F: GGATCCATGGAGCAAAATTTGTCCCT | 用于原核表达载体构建 |
| R: AAGCTTAAACTTAAAACCGTCGTCAA | ||
| TT2-VIGS | F: GGACTAGTCTTCATAAACTCCTTGGGAAC | 用于基因沉默载体构建 |
| R: TTGGCGCGCCCCACTCAAAGTTCCGGGTATC | ||
| TT2-qRT | F: TTCCATCCATAGCAGGTTTA | 用于实时荧光定量PCR |
| R: CCTGTTCCCAAGGAGTTTAT | ||
| GhUBQ7-qRT | F: GAAGGCATTCCACCTGACCAAC | 用于实时荧光定量PCR |
| R: CTTGACCTTCTTCTTGTGCTTG |
| [1] | Su QS, Wang S, Sun YQ, Mei J, Ke LP. Advances in biosynthesis and regulation of plant proanthocyanidins. Chin J Cell Biol, 2021, 43(1): 219-229. |
| 苏全胜, 王爽, 孙玉强, 梅俊, 柯丽萍. 植物原花青素生物合成及调控研究进展. 中国细胞生物学学报, 2021, 43(1): 219-229. | |
| [2] |
Liu WX, Feng Y, Yu SH, Fan ZQ, Li XL, Li JY, Yin HF. The flavonoid biosynthesis network in plants. Int J Mol Sci, 2021, 22(23): 12824.
doi: 10.3390/ijms222312824 |
| [3] |
Dixon RA, Sarnala S. Proanthocyanidin biosynthesis-a matter of protection. Plant Physiol, 2020, 184(2): 579-591.
doi: 10.1104/pp.20.00973 |
| [4] |
Xie DY, Sharma SB, Paiva NL, Ferreira D, Dixon RA. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science, 2003, 299(5605): 396-399.
doi: 10.1126/science.1078540 |
| [5] |
Sharma SB, Dixon RA. Metabolic engineering of proanthocyanidins by ectopic expression of transcription factors in Arabidopsis thaliana. Plant J, 2005, 44(1): 62-75.
doi: 10.1111/j.1365-313X.2005.02510.x |
| [6] |
Vale M, Rodrigues J, Badim H, Gerós H, Conde A. Exogenous application of non-mature miRNA-encoded miPEP164c inhibits proanthocyanidin synthesis and stimulates anthocyanin accumulation in grape berry cells. Front Plant Sci, 2021, 12: 706679.
doi: 10.3389/fpls.2021.706679 |
| [7] |
Gil-Muñoz F, Sánchez-Navarro JA, Besada C, Salvador A, Badenes ML, Del Mar Naval M, Ríos G. MBW complexes impinge on anthocyanidin reductase gene regulation for proanthocyanidin biosynthesis in persimmon fruit. Sci Rep, 2020, 10(1): 3543.
doi: 10.1038/s41598-020-60635-w |
| [8] |
Gao YF, Liu JK, Chen YF, Tang H, Wang Y, He YM, Ou YB, Sun XC, Wang SH, Yao YN. Tomato SlAN11 regulates flavonoid biosynthesis and seed dormancy by interaction with bHLH proteins but not with MYB proteins. Hortic Res, 2018, 5: 27.
doi: 10.1038/s41438-018-0032-3 |
| [9] | Yuan JL, Zheng HL, Liang XL, Mei J, Yu DL, Sun YQ, Ke LP. Influence of anthocyanin biosynthesis on leaf and fiber color of Gossypium hirsutum L. Sci Agric Sin, 2021, 54(9): 1847-1855. |
| 袁景丽, 郑红丽, 梁先利, 梅俊, 余东亮, 孙玉强, 柯丽萍. 花青素代谢对陆地棉叶片和纤维色泽呈现的影响. 中国农业科学, 2021, 54(9): 1847-1855. | |
| [10] | Du QL, Fang YP, Jiang JM, Li XY, Xie X. Cloning, expression analysis and prokaryotic expression of rice OsHDAC15 gene. Plant Physiol J, 2021, 57(3): 558-568. |
| 杜巧丽, 方远鹏, 蒋君梅, 李向阳, 谢鑫. 水稻OsHDAC15基因的克隆、表达分析及原核表达. 植物生理学报, 2021, 57(3): 558-568. | |
| [11] |
Wang JL, Chen SS, Jiang N, Li N, Wang XY, Li ZP, Li X, Liu HT, Li L, Yang Y, Ni T, Yu CY, Ma JB, Zheng BL, Ren GD.Spliceosome disassembly factors ILP1 and NTR1 promote miRNA biogenesis in Arabidopsis thaliana. Nucleic Acids Res, 2019, 47(15): 7886-7900.
doi: 10.1093/nar/gkz526 |
| [12] |
Mei J, Jiang N, Ren GD. The F-box protein HAWAIIAN SKIRT is required for mimicry target-induced microRNA degradation in Arabidopsis. J Integr Plant Biol, 2019, 61(11): 1121-1127.
doi: 10.1111/jipb.12761 |
| [13] |
Gao JF, Shen L, Yuan JL, Zheng HL, Su QS, Yang WG, Zhang LQ, Nnaemeka VE, Sun J, Ke LP, Sun YQ. Functional analysis of GhCHS, GhANR and GhLAR in colored fiber formation of Gossypium hirsutum L. BMC Plant Biol, 2019, 19(1): 455.
doi: 10.1186/s12870-019-2065-7 |
| [14] |
Yang SC, Zhang M, Xu LQ, Luo ZR, Zhang QL. MiR858b inhibits proanthocyanidin accumulation by the repression of DkMYB19 and DkMYB20 in persimmon. Front Plant Sci, 2020, 11: 576378.
doi: 10.3389/fpls.2020.576378 |
| [15] |
Wang LJ, Ran LY, Hou YS, Tian QY, Li CF, Liu R, Fan D, Luo KM. The transcription factor MYB115contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. New Phytol, 2017, 215(1): 351-367.
doi: 10.1111/nph.14569 |
| [16] |
Shen YX, Sun TT, Pan Q, Anupol N, Chen H, Shi JW, Liu F, Deqiang D, Wang CQ, Zhao J, Yang SH, Wang CY, Liu JH, Bao MZ, Ning GG.RrMYB5- and RrMYB10-regulated flavonoid biosynthesis plays a pivotal role in feedback loop responding to wounding and oxidation in Rosa rugosa. Plant Biotechnol J, 2019, 17(11): 2078-2095.
doi: 10.1111/pbi.13123 |
| [17] | Lu N, Roldan M, Dixon RA. Characterization of two TT2-type MYB transcription factors regulating proanthocyanidin biosynthesis in tetraploid cotton, Gossypium hirsutum. Planta, 2017, 246(2): 323-335. |
| [18] |
Sun J, Sun YQ, Zhu QH. Breeding next-generation naturally colored cotton. Trends Plant Sci, 2021, 26(6): 539-542.
doi: 10.1016/j.tplants.2021.03.007 |
| [19] |
Liu HF, Luo C, Song W, Shen HT, Li GL, He ZG, Chen WG, Cao YY, Huang F, Tang SW, Hong P, Zhao EF, Zhu JB, He DJ, Wang SM, Huo GY, Liu HL. Flavonoid biosynthesis controls fiber color in naturally colored cotton. Peer J, 2018, 6: e4537.
doi: 10.7717/peerj.4537 |
| [20] |
Hinchliffe DJ, Condon BD, Thyssen G, Naoumkina M, Madison CA, Reynolds M, Delhom CD, Fang DD, Li P, McCarty J. The GhTT2_A07gene is linked to the brown colour and natural flame retardancy phenotypes of Lc1 cotton (Gossypium hirsutum L.) fibres. J Exp Bot, 2016, 67(18): 5461-5471.
pmid: 27567364 |
| [21] |
Yan Q, Wang Y, Li Q, Zhang ZS, Ding H, Zhang Y, Liu HS, Luo M, Liu DX, Song W, Liu HF, Yao D, Ouyang XF, Li YH, Li X, Pei Y, Xiao YH. Up-regulation of GhTT2-3A in cotton fibres during secondary wall thickening results in brown fibres with improved quality. Plant Biotechnol J, 2018, 16(10): 1735-1747.
doi: 10.1111/pbi.12910 |
| [1] | Xiaohua Hao, Shuang Hu, Dan Zhao, Lianfu Tian, Zijing Xie, Sha Wu, Wenli Hu, Han Lei, Dongping Li. OsGA3ox genes regulate rice fertility and plant height by synthesizing diverse active GA [J]. Hereditas(Beijing), 2023, 45(9): 845-855. |
| [2] | Fengyu Sun, Qianghua Xu. Research progress of microRNAs involved in hematopoiesis [J]. Hereditas(Beijing), 2022, 44(9): 756-771. |
| [3] | Menggang Lv, Aijia Liu, Qingwei Li, Peng Su. Progress on the origin, function and evolutionary mechanism of RHR transcription factor family [J]. Hereditas(Beijing), 2021, 43(3): 215-225. |
| [4] | Xiaofen Qiu, Dong’e Tang, Haiyan Yu, Qiuyan Liao, Zhiyang Hu, Jun Zhou, Xin Zhao, Huiyan He, Zhuojian Liang, Chengming Xu, Ming Yang, Yong Dai. Analysis of transcription factors in accessible open chromatin in the 18-trisomy syndrome based on single cell ATAC sequencing technique [J]. Hereditas(Beijing), 2021, 43(1): 74-83. |
| [5] | Xiaomei Tuo, Dongli Zhu, Xiaofeng Chen, Yu Rong, Yan Guo, Tielin Yang. The osteoporosis susceptible SNP rs4325274 remotely regulates the SOX6 gene through enhancers [J]. Hereditas(Beijing), 2020, 42(9): 889-897. |
| [6] | Jie Wu, Jianping Quan, Yong Ye, ZhenFang Wu, Jie Yang, Ming Yang, Enqin Zheng. Advances in assay for transposase-accessible chromatin with high-throughput sequencing [J]. Hereditas(Beijing), 2020, 42(4): 333-346. |
| [7] | Taotao Wang, Yong Yang, Wei Wei, Chentao Lin, Liuyin Ma. Identification and expression analyses of the NAC transcription factor family in Spartina alterniflora [J]. Hereditas(Beijing), 2020, 42(2): 194-211. |
| [8] | Zhaoqing Sun, Bo Yan. The roles and regulation mechanism of transcription factor GATA6 in cardiovascular diseases [J]. Hereditas(Beijing), 2019, 41(5): 375-383. |
| [9] | Haoqiang Yu,Fuai Sun,Wenqi Feng,Fengzhong Lu,Wanchen Li,Fengling Fu. The BES1/BZR1 transcription factors regulate growth, development and stress resistance in plants [J]. Hereditas(Beijing), 2019, 41(3): 206-214. |
| [10] | Junyi Ju,Quan Zhao. Regulation of γ-globin gene expression and its clinical applications [J]. Hereditas(Beijing), 2018, 40(6): 429-444. |
| [11] | Junxia Zou, Ke Chen. Roles and molecular mechanisms of hypoxia-inducible factors in renal cell carcinoma [J]. Hereditas(Beijing), 2018, 40(5): 341-356. |
| [12] | Qingqian Ding,Xiaoting Wang,Liqin Hu,Xin Qi,Linhao Ge,Weiya XU,Zhaoshi Xu,Yongbin Zhou,Guanqing Jia,Xianmin Diao,Donghong Min,Youzhi Ma,Ming Chen. MYB-like transcription factor SiMYB42 from foxtail millet (Setaria italica L.) enhances Arabidopsis tolerance to low-nitrogen stress [J]. Hereditas(Beijing), 2018, 40(4): 327-338. |
| [13] | Lan Ren,Rudan Xiao,Qian Zhang,Xiaomin Lou,Zhaojun Zhang,Xiangdong Fang. Synergistic regulation of the erythroid differentiation of K562 cells by KLF1 and KLF9 [J]. Hereditas(Beijing), 2018, 40(11): 998-1006. |
| [14] | Ling Zhang, Jianbo He. Progress of GATA6 in liver development [J]. Hereditas(Beijing), 2018, 40(1): 22-32. |
| [15] | Lei Xu,Wen Chen,Guoyang Si,Yiyuan Huang,Yi Lin,Yongping Cai,Junshan Gao. Genome-wide analysis of the GST gene family in Gossypium hirsutum L. [J]. Hereditas(Beijing), 2017, 39(8): 737-752. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||