Hereditas(Beijing) ›› 2020, Vol. 42 ›› Issue (4): 333-346.doi: 10.16288/j.yczz.19-279
• Review • Next Articles
Advances in assay for transposase-accessible chromatin with high-throughput sequencing
Jie Wu1, Jianping Quan1, Yong Ye1, ZhenFang Wu1, Jie Yang1, Ming Yang2( ), Enqin Zheng1(
), Enqin Zheng1( )
)
- 1. National Engineering Research Center for Breeding Swine Industry, College of Animal Science, South China Agricultural University, Guangzhou 510642, China
2. College of Animal Science and Technology, Zhongkai University of Agriculture and Engineering, Guangzhou 510225, China
-
Received:2019-11-14Revised:2020-01-29Online:2020-04-20Published:2020-02-26 -
Contact:Yang Ming,Zheng Enqin E-mail:yangming@zhku.edu.cn;eqzheng@scau.edu.cn -
Supported by:Supported by Guangdong YangFan Innovative and Entrepreneurial Research Team Program No(2016YT03H062);Guangdong Modern Agricultural Industry Technology System Pig Innovation Team Project No(2019KJ126);Guangdong Natural Science Foundation(2017A030313213)
Cite this article
Jie Wu, Jianping Quan, Yong Ye, ZhenFang Wu, Jie Yang, Ming Yang, Enqin Zheng. Advances in assay for transposase-accessible chromatin with high-throughput sequencing[J]. Hereditas(Beijing), 2020, 42(4): 333-346.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Introduction of five chromatin accessibility assays"
| 技术 | 细胞类型及数量 | 获取方式 | 目的 | 特点 | 参考文献 |
|---|---|---|---|---|---|
| DNase-seq | 需要1,000,000~ 10,000,000的任何 细胞类型 | DNase I 酶切 | 获取染色质开放信息 | (1)比传统方法操作更简便; (2)细胞需要量大; (3)酶最优酶切浓度确定过程繁琐; (4)样品制备过程复杂且耗时 | [20,24] |
| MNase-seq | 需要1,000,000~ 10,000,000的 任何细胞类型 | 微球菌 核酸酶酶切 | 绘制核小体图谱以间 接探测染色质可及性 | (1)操作简单,后期数据处理方便; (2)细胞样本需要量大; (3)酶浓度和切割温度难以确定 | [26,27] |
| FAIRE-seq | 需要100,000~ 10,000,000的任 何细胞类型 | 超声波 物理断裂 | 获取染色质开放信息 | (1)不用酶切、不需要分离出细胞核; (2)没有序列切割特异性; (3)细胞需要量大; (4)甲醛最佳交联程度难以确定 | [21,24,31] |
| NOMe-seq | 至少1,000,000 的任何细胞类型 | 甲基化修饰 | 获得内源DNA甲基化 的信息并定位核小体 | (1)不需要使DNA断裂,不会产生富集偏差; (2)同时获得含GpC和CpG二核苷酸的信息; (3)细胞需要量大 | [22,34] |
| ATAC-seq | 500~50,000个 新鲜分离的细胞 | Tn5转座 酶酶切 | 获取染色质可及性、 转录因子结合以及 核小体定位信息 | (1)过程简便、效率高; (2)数据分析工具不够成熟; (3)线粒体、叶绿体中的DNA污染; (4)冷冻组织细胞DNA提取效率低; (5) DNA片段损失过多 | [36~38] |
| [1] | Kornberg RD . Chromatin structure: a repeating unit of histones and DNA. Science, 1974,184(4139):868-871. |
| [2] | Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A . Structure of the nucleosome core particle at 7 A resolution. Nature, 1984,311(5986):532-537. |
| [3] | Zhou YB, Gerchman SE, Ramakrishnan V, Travers A, Muyldermans S . Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature, 1998,395(6700):402-405. |
| [4] | Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA . The accessible chromatin landscape of the human genome. Nature, 2012,489(7414):75-82. |
| [5] | Poirier MG, Bussiek M, Langowski J, Widom J . Spontaneous access to DNA target sites in folded chromatin fibers. J Mol Biol, 2008,379(4):772-786. |
| [6] | Fedor MJ . Chromatin structure and gene expression. Curr Opin Cell Biol, 1996,4(18):9384-9388. |
| [7] | John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA . Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet, 2011,43(3):264-268. |
| [8] | Bell O, Tiwari VK, Thomä NH, Schübeler D . Determinants and dynamics of genome accessibility. Nat Rev Genet, 2011,12(8):554-564. |
| [9] | Kouzarides T . Chromatin modifications and their function. Cell, 2007,128(4):693-705. |
| [10] | Jiang C, Pugh BF . Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet, 2009,10(3):161-172. |
| [11] | Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang ZB, Wei G, Zhao KJ . Dynamic regulation of nucleosome positioning in the human genome. Cell, 2008,132(5):887-898. |
| [12] | Hewish DR, Burgoyne LA . Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun, 1973,52(2):504-510. |
| [13] | Scott WA, Wigmore DJ . Sites in simian virus 40 chromatin which are preferentially cleaved by endonucleases. Cell, 1978,15(4):1511-1518. |
| [14] | Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SCR . The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell, 1979,16(4):797-806. |
| [15] | Stalder J, Larsen A, Engel JD, Dolan M, Groudine M, Weintraub H . Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNase I. Cell, 1980,20(2):451-460. |
| [16] | Mcghee JD, Wood WI, Dolan M, Engel JD, Felsenfeld G . A 200 base pair region at the 5’ end of the chicken adult β-globin gene is accessible to nuclease digestion. Cell, 1981,27(1, Part 2):45-55. |
| [17] | Gross DS, Garrard WT . Nuclease hypersensitive sites in chromatin. Annu Rev Biochem, 2003,57(57):159-197. |
| [18] | Wu C . The 5’ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature, 1980,286(5776):854-860. |
| [19] | Crawford GE, Holt IE, Mullikin JC, Tai D, Blakesley R, Bouffard G, Young A, Masiello C, Green ED, Wolfsberg TG, Collins FS . Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc Natl Acad Sci USA, 2004,101(4):992-997. |
| [20] | Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE . High-resolution mapping and characterization of open chromatin across the genome. Cell, 2008,132(2):311-322. |
| [21] | Giresi PG, Kim J, Mcdaniell RM, Iyer VR, Lieb JD . FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res, 2007,17(6):877-885. |
| [22] | Kelly TK, Liu YP, Lay FD, Liang GN, Berman BP, Jones PA . Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res, 2012,22(12):2497-2506. |
| [23] | He HH, Meyer CA, Hu SS, Chen MW, Zang C, Liu Y, Rao PK, Fei T, Xu H, Long H, Liu XS, Brown M . Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods, 2014,11(1):73-78. |
| [24] | Tsompana M, Buck MJ . Chromatin accessibility: a window into the genome. Epigenetics Chromatin, 2014,7(1):33. |
| [25] | Cumbie JS, Filichkin SA, Megraw M . Improved DNase-seq protocol facilitates high resolution mapping of DNase I hypersensitive sites in roots in Arabidopsis thaliana. Plant Methods, 2015,11(1):42. |
| [26] | Rizzo JM, Sinha S . Analyzing the global chromatin structure of keratinocytes by MNase-seq. Methods Mol Biol, 2014,1195:49-59. |
| [27] | Telford DJ, Stewart BW . Micrococcal nuclease: its specificity and use for chromatin analysis. Int J Biochem, 1989,21(2):127-137. |
| [28] | Chung HR, Dunkel I, Heise F, Linke C, Krobitsch S, Ehrenhofer-Murray AE, Sperling SR, Vingron M . The effect of micrococcal nuclease digestion on nucleosome positioning data. PLoS One, 2010,5(12):e15754. |
| [29] | Clark DJ. Nucleosome Positioning , Nucleosome spacing and the nucleosome code. J Biomol Struct Dyn, 2010,27(6):781-793. |
| [30] | Zentner GE, Henikoff S . Surveying the epigenomic landscape, one base at a time. Genome Biol, 2012,13(10):250. |
| [31] | Kumar V, Muratani M, Rayan NA, Kraus P, Lufkin T, Ng HH, Prabhakar S . Uniform, optimal signal processing of mapped deep-sequencing data. Nat Biotechnol, 2013,31(7):615-622. |
| [32] | Simon JM, Giresi PG, Davis IJ, Lieb JD . Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc, 2012,7(2):256-267. |
| [33] | Auerbach RK, Euskirchen G, Rozowsky J, Lamarre- Vincent N, Moqtaderi Z, Lefrancois P, Struhl K, Gerstein M, Snyder M . Mapping accessible chromatin regions using Sono-Seq. Proc Natl Acad Sci USA, 2009,106(35):14926-14931. |
| [34] | Rhie SK, Schreiner S, Farnham PJ . Defining regulatory elements in the human genome using nucleosome occupancy and methylome sequencing (NOMe-Seq). Methods Mol Biol, 2018,1766:209-229. |
| [35] | Klemm SL, Shipony Z, Greenleaf WJ . Chromatin accessibility and the regulatory epigenome. Nat Rev Genet, 2019,20(4):207-220. |
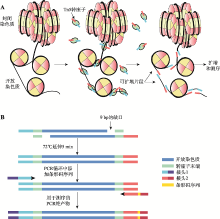
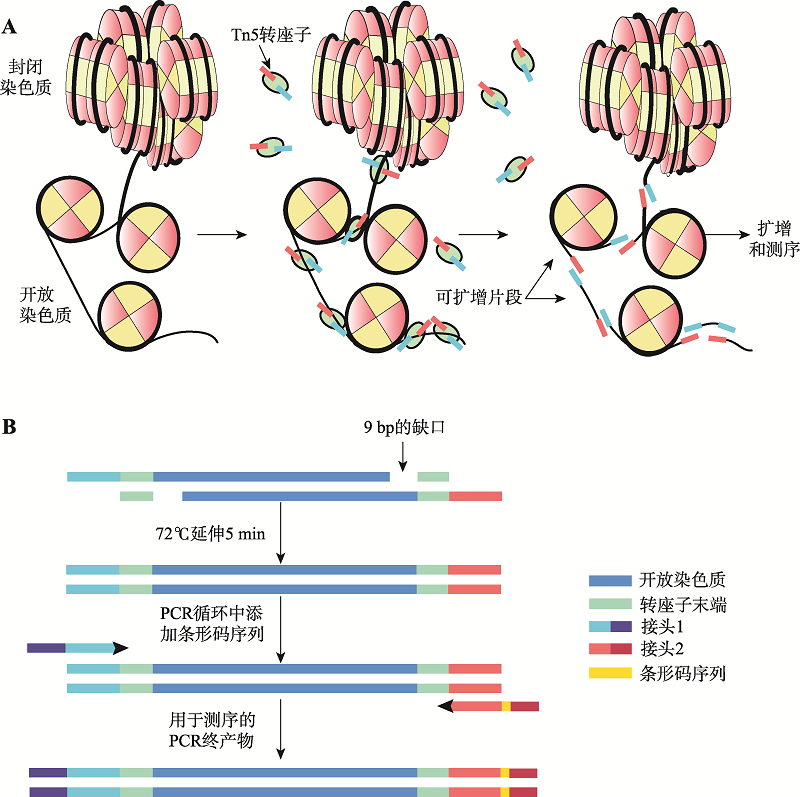
| [36] | Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ . Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Met, 2013,10(12):1213-1218. |
| [37] | Wu JY, Huang B, Chen H, Yin QZ, Liu Y, Xiang YL, Zhang BJ, Liu BF, Wang QJ, Xia WK, Li WZ, Li YY, Ma J, Peng X, Zheng H, Ming J, Zhang WH, Zhang J, Tian G, Xu F, Chang Z, Na J, Yang XR, Xie W . The landscape of accessible chromatin in mammalian preimplantation embryos. Nature, 2016,534(7609):652-657. |
| [38] | Buenrostro JD, Wu B, Chang HY, Greenleaf WJ . ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol, 2015,109:21-29. |
| [39] | Mcclintock B . The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA, 1950,36(6):344-355. |
| [40] | Bucher E, Reinders J, Mirouze M . Epigenetic control of transposon transcription and mobility in Arabidopsis. Curr Opin Plant Biol, 2012,15(5):503-510. |
| [41] | Huang CR, Burns KH, Boeke JD . Active transposition in genomes. Annu Rev Genet, 2012,46:651-675. |
| [42] | Berg DE, Davies J, Allet B, Rochaix JD . Transposition of R factor genes to bacteriophage lambda. Proc Natl Acad Sci USA, 1975,72(9):3628-3632. |
| [43] | Rothstein SJ, Jorgensen RA, Postle K, Reznikoff WS . The inverted repeats of Tn5 are functionally different. Cell, 1980,19(3):795-805. |
| [44] | Lovell S, Goryshin IY, Reznikoff WR, Rayment I . Two-metal active site binding of a Tn5 transposase synaptic complex. Nat Struct Biol, 2002,9(4):278-281. |
| [45] | Auerswald EA, Ludwig G, Schaller H. Structural analysis of Tn5. Cold Spring Harb Symp Quant Biol, 1981, 45 Pt1:107. |
| [46] | Berg DE . Julian Davies and the discovery of kanamycin resistance transposon Tn5. J Antibiot, 2017,70(4):339-346. |
| [47] | Reznikoff WS . The TN5 transposon. Annu Rev Microbiol, 1993,47(1):945-963. |
| [48] | Reznikoff WS, Bhasin A, Davies DR, Goryshin IY, Mahnke LA, Naumann T, Rayment I, Steiniger-White M, Twining SS . Tn5: A molecular window on transposition. Biochem Biophys Res Commun, 1999,266(3):729-734. |
| [49] | Zhou M, Reznikoff WS . Tn5 transposase mutants that alter DNA binding specificity. J Mol Biol, 1997,271(3):362-373. |
| [50] | York D, Reznikoff WS . DNA binding and phasing analyses of Tn5 transposase and a monomeric variant. Nucleic Acids Res, 1997,25(11):2153-2160. |
| [51] | Steiniger-White M, Reznikoff WS . The C-terminal alpha helix of Tn5 transposase is required for synaptic complex formation. J Biol Chem, 2000,275(30):23127-23133. |
| [52] | Bhasin A, Goryshin IY, Steiniger-White M, York D, Reznikoff WS . Characterization of a Tn5 pre-cleavage synaptic complex. J Mol Biol, 2000,302(1):49-63. |
| [53] | Kale SB, Landree MA, Roth DB . Conditional RAG-1 mutants block the hairpin formation step of V(D)J recombination. Mol Cell Biol, 2001,21(2):459-466. |
| [54] | Naumann TA, Reznikoff WS . Trans catalysis in Tn5 transposition. Proc Natl Acad Sci USA, 2000,97(16):8944-8949. |
| [55] | Lisa AMB, Goryshin IY, Reznikoff WS . A mechanism for Tn5 inhibition. J Biol Chem, 1999,274(1):86. |
| [56] | Bhasin A, Goryshin IY, Reznikoff WS . Hairpin formation in Tn5 transposition. J Biol Chem, 1999,274(52):37021-37029. |
| [57] | Mizuuchi K, Adzuma K . Inversion of the phosphate chirality at the target site of Mu DNA strand transfer: Evidence for a one-step transesterification mechanism. Cell, 1991,66(1):129-140. |
| [58] | Mizuuchi K . Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem, 1992,61:1011-1051. |
| [59] | Crellin P, Chalmers R . Protein-DNA contacts and conformational changes in the Tn10 transpososome during assembly and activation for cleavage. EMBO J, 2001,20(14):3882-3891. |
| [60] | Goryshin IY, Reznikoff WS . Tn5 in vitro transposition. J Biol Chem, 1998,273(13):7367-7374. |
| [61] | Adey A, Morrison HG, Asan , Xun X, Kitzman JO, Turner EH, Stackhouse B, Mackenzie AP, Caruccio NC, Zhang X, Shendure J . Rapid, low-input, low-bias construction of shotgun fragment libraries by high- density in vitro transposition. Genome Biol, 2010,11(12):R119. |
| [62] | Reznikoff WS . Transposon Tn5. Annu Rev Genet, 2008,42:269-286. |
| [63] | Davies DR, Mahnke Braam L, Reznikoff WS, Rayment I . The three-dimensional structure of a Tn5 transposase- related protein determined to 2.9-A resolution. J Biol Chem, 1999,274(17):11904-11913. |
| [64] | Peterson G, Reznikoff W . Tn5 transposase active site mutations suggest position of donor backbone DNA in synaptic complex. J Biol Chem, 2003,278(3):1904-1909. |
| [65] | Reznikoff WS . Tn5 as a model for understanding DNA transposition. Mol Microbiol, 2003,47(5):1199-1206. |
| [66] | Sakamoto H, Thiberge S, Akerman S, Janse CJ, Carvalho TG, Ménard R . Towards systematic identification of Plasmodium essential genes by transposon shuttle mutagenesis. Nucleic Acids Res, 2005,33(20):e174. |
| [67] | Caruccio N . Preparation of next-generation sequencing libraries using Nextera TM technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition . Methods Mol Biol, 2011,733:241-255. |
| [68] | Chen CY, Xing D, Tan LZ, Li H, Zhou GY, Huang L, Xie XS . Single-cell whole-genome analyses by linear amplification via transposon insertion (LIANTI). Science, 2017,356(6334):189-194. |
| [69] | Chen XQ, Shen Y, Draper W, Buenrostro JD, Litzenburger U, Cho SW, Satpathy AT, Carter AC, Ghosh RP, East-Seletsky A, Doudna JA, Greenleaf WJ, Liphardt JT, Chang HY . ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods, 2016,13(12):1013-1020. |
| [70] | Buenrostro JD Giresi PG Zaba LC, Chang HY, Greenleat W . Transposition of native chromatin for multimodal regulatory analysis and personal epigenomics. Nat Methods, 2013,10(12):1213-1218. |
| [71] | Lu Z, Hofmeister BT, Vollmers C, Dubois RM, Schmitz RJ . Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res, 2017,45(6):e41. |
| [72] | Deal RB, Henikoff S . A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell, 2010,18(6):1030-1040. |
| [73] | Maher KA, Bajic M, Kajala K, Reynoso M, Pauluzzi G, West DA, Zumstein K, Woodhouse M, Bubb K, Dorrity MW, Queitsch C, Bailey-Serres J, Sinha N, Brady SM, Deal RB . Profiling of accessible chromatin regions across multiple plant species and cell types reveals common gene regulatory principles and new control modules. Plant Cell, 2018,30(1):15-36. |
| [74] | Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, Kathiria A, Cho SW, Mumbach MR, Carter AC, Kasowski M, Orloff LA, Risca VI, Kundaje A, Khavari PA, Montine TJ, Greenleaf WJ, Chang HY . An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods, 2017,14(10):959-962. |
| [75] | Sos BC, Fung HL, Gao DR, Osothprarop TF, Kia A, He MM, Zhang K . Characterization of chromatin accessibility with a transposome hypersensitive sites sequencing (THS-seq) assay. Genome Biol, 2016,17(1):20. |
| [76] | Tan L, Ke Z, Tombline G, Macoretta N, Hayes K, Tian X, Lv R, Ablaeva J, Gilbert M, Bhanu NV, Yuan Z F, Garcia BA, Shi YG, Shi Y, Seluanov A, Gorbunova V . Naked mole rat cells have a stable epigenome that resists iPSC reprogramming. Stem Cell Reports, 2017,9(5):1721-1734. |
| [77] | Scharer CD, Blalock EL, Barwick BG, Haines RR, Wei C, Sanz I, Boss JM . ATAC-seq on biobanked specimens defines a unique chromatin accessibility structure in naïve SLE B cells. Sci Rep, 2016,6(1):27030. |
| [78] | Davie K, Jacobs J, Atkins M, Potier D, Christiaens V, Halder G, Aerts S . Discovery of transcription factors and regulatory regions driving in vivo tumor development by ATAC-seq and FAIRE-seq open chromatin profiling. PLoS Genet, 2015,11(2):e1004994. |
| [79] | Liu YJ, Zhang F, Liu HD, Sun X . The application of next-generation sequencing techniques in studying transcriptional regulation in embryonic stem cells. Hereditas(Beijing), 2017,39(8):717-725. |
| 刘亚军, 张峰, 刘宏德, 孙啸 . 下一代测序技术在干细胞转录调控研究中的应用. 遗传, 2017,39(8):717-725. | |
| [80] | Quillien A, Abdalla M, Yu J, Ou JH, Zhu LJ, Lawson ND . Robust identification of developmentally active endothelial enhancers in zebrafish using FANS-assisted ATAC-Seq. Cell Rep, 2017,20(3):709-720. |
| [81] | Ackermann AM, Wang ZP, Schug J, Naji A, Kaestner KH . Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol Metab, 2016,5(3):233-244. |
| [82] | Garcia E, Hayden A, Birts C, Britton E, Cowie A, Pickard K, Mellone M, Choh C, Derouet M, Duriez P, Noble F, White MJ, Primrose JN, Strefford JC, Rose-Zerilli M, Thomas GJ, Ang Y, Sharrocks AD, Fitzgerald RC, Underwood TJ . Authentication and characterisation of a new oesophageal adenocarcinoma cell line: MFD-1. Sci Rep, 2016,6:32417. |
| [83] | Ho YT, Shimbo T, Wijaya E, Ouchi Y, Takaki E, Yamamoto R, Kikuchi Y, Kaneda Y, Tamai K . Chromatin accessibility identifies diversity in mesenchymal stem cells from different tissue origins. Sci Rep, 2018,8(1):17765. |
| [84] | Nelson AC, Mould AW, Bikoff EK, Robertson EJ . Mapping the chromatin landscape and Blimp1 transcriptional targets that regulate trophoblast differentiation. Sci Rep, 2017,7(1):6715-6793. |
| [85] | Scott-Browne JP, López-Moyado IF, Trifari S, Wong V, Chavez L, Rao A, Pereira RM . Dynamic changes in chromatin accessibility occur in CD8 + T cells responding to viral infection . Immunity, 2016,45(6):1327-1340. |
| [86] | Starks RR, Biswas A, Jain A, Tuteja G . Combined analysis of dissimilar promoter accessibility and gene expression profiles identifies tissue-specific genes and actively repressed networks. Epigenetics Chromatin, 2019,12(1):16. |
| [87] | Rajbhandari P, Thomas B J, Feng A, Hong C, Wang JX, Vergnes L, Sallam T, Wang B, Sandhu J, Seldin M M, Lusis AJ, Fong LG, Katz M, Lee R, Young SG, Reue K, Smale ST, Tontonoz P. IL-10 signaling remodels adipose chromatin architecture to limit thermogenesis and energy expenditure. Cell, 2018, 172(1-2): 218-233. e17. |
| [88] | Denny SK, Yang D, Chuang CH, Brady JJ, Lim JS, Grüner BM, Chiou S, Schep AN, Baral J, Hamard C, Antoine M, Wislez M, Kong CS, Connolly AJ, Park K, Sage J, Greenleaf WJ, Winslow MM . Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell, 2016,166(2):328-342. |
| [89] | Wang MJ, Wang PC, Lin M, Ye ZX, Li GL, Tu LL, Shen C, Li JY, Yang QY, Zhang XL . Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nat Plants, 2018,4(2):90-97. |
| [90] | Mas G, Blanco E, Ballaré C, Sansó M, Spill YG, Hu D, Aoi Y, Le Dily F, Shilatifard A, Marti-Renom MA, Di Croce L . Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat Genet, 2018,50(10):1452-1462. |
| [91] | Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, Haugen E, Heimfeld S, Murry CE, Akey JM, Stamatoyannopoulos JA . Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell, 2013,154(4):888-903. |
| [92] | Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ . Single-cell chromatin accessibility reveals principles of regulatory variation. Nature, 2015,523(7561):486-490. |
| [93] | Liu LQ, Liu YC, Quintero A, Wu L, Yuan Y, Wang MY, Cheng MN, Leng LZ, Xu LQ, Dong GY, Li R, Liu Y, Wei XY, Xu JS, Chen XW, Lu HR, Chen DS, Wang QL, Zhou Q, Lin XX, Li GB, Liu SP, Wang Q, Wang HR, Fink JL, Gao ZL, Liu X, Hou Y, Zhu SD, Yang HM, Ye YM, Lin G, Chen F, Herrmann C, Eils R, Shang ZC, Xu X . Deconvolution of single-cell multi-omics layers reveals regulatory heterogeneity. Nat Commun, 2019,10(1):470. |
| [94] | Cusanovich DA, Hill AJ, Aghamirzaie D, Daza RM, Pliner HA, Berletch JB, Filippova GN, Huang X, Christiansen L, Dewitt WS, Lee C, Regalado SG, Read DF, Steemers FJ, Disteche CM, Trapnell C, Shendure J . A single-cell atlas of in vivo mammalian chromatin accessibility. Cell, 2018,174(5):1309-1324. |
| [95] | Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, Steemers FJ, Trapnell C, Shendure J . Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science, 2015,348(6237):910-914. |
| [96] | Zamanighomi M, Lin ZL, Daley T, Chen X, Duren Z, Schep A, Greenleaf WJ, Wong WH . Unsupervised clustering and epigenetic classification of single cells. Nat Commun, 2018,9(1):2410. |
| [97] | Schep AN, Wu BJ, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods, 2017,14(10):975-978. |
| [98] | Baker SM, Rogerson C, Hayes A, Sharrocks AD, Rattray M . Classifying cells with Scasat, a single-cell ATAC-seq analysis tool. Nucleic Acids Res, 2019,47(2):e10. |
| [99] | Preissl S, Fang RX, Huang H, Zhao Y, Raviram R, Gorkin DU, Zhang YX, Sos BC, Afzal V, Dickel DE, Kuan S, Visel A, Pennacchio LA, Zhang K, Ren B . Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat Neurosci, 2018,21(3):432-439. |
| [100] | Cusanovich DA, Reddington JP, Garfield DA, Daza RM, Aghamirzaie D, Marco-Ferreres R, Pliner HA, Christiansen L, Qiu X, Steemers FJ, Trapnell C, Shendure J, Furlong EEM . The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature, 2018,555(7697):538-542. |
| [101] | Buenrostro JD, Corces MR, Lareau CA, Wu B, Schep AN, Aryee MJ, Majeti R, Chang HY, Greenleaf WJ . Integrated single-cell analysis maps the continuous regulatory landscape of human hematopoietic differentiation. Cell, 2018,173(6):1535-1548. |
| [102] | Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM, Noble WS, Duan Z, Shendure J . Massively multiplex single-cell Hi-C. Nature Methods, 2017,14(3):263-266. |
| [1] | Fengyu Sun, Qianghua Xu. Research progress of microRNAs involved in hematopoiesis [J]. Hereditas(Beijing), 2022, 44(9): 756-771. |
| [2] | Rongrong Mu, Qingqing Niu, Yuqiang Sun, Jun Mei, Meng Miao. Cloning and characterization of the MYB transcription factor gene GhTT2 in Gossypium hirsutum [J]. Hereditas(Beijing), 2022, 44(8): 720-728. |
| [3] | Hui Qu, Yi Liu, Yawen Chen, Hui Wang. Alteration of imprinted genes and offspring organ development caused by environmental factors [J]. Hereditas(Beijing), 2022, 44(2): 107-116. |
| [4] | Yangjinghui Zhang, Peiyao Chang, Zishu Yang, Yuhang Xue, Xueqi Li, Yang Zhang. Advances in epigenetic modification affecting anthocyanin synthesis [J]. Hereditas(Beijing), 2022, 44(12): 1117-1127. |
| [5] | Qingwen Zhao, Dongning Pan. Progress on the epigenetic regulation of adipose tissue thermogenesis [J]. Hereditas(Beijing), 2022, 44(10): 867-880. |
| [6] | Wang Ya'nan, Tao Xu, Wanpeng Wang, Qingzhu Zhang, Xie Li'nan. Role of epigenetic modifications in the development of crops essential traits [J]. Hereditas(Beijing), 2021, 43(9): 858-879. |
| [7] | Menggang Lv, Aijia Liu, Qingwei Li, Peng Su. Progress on the origin, function and evolutionary mechanism of RHR transcription factor family [J]. Hereditas(Beijing), 2021, 43(3): 215-225. |
| [8] | Xiaofen Qiu, Dong’e Tang, Haiyan Yu, Qiuyan Liao, Zhiyang Hu, Jun Zhou, Xin Zhao, Huiyan He, Zhuojian Liang, Chengming Xu, Ming Yang, Yong Dai. Analysis of transcription factors in accessible open chromatin in the 18-trisomy syndrome based on single cell ATAC sequencing technique [J]. Hereditas(Beijing), 2021, 43(1): 74-83. |
| [9] | Xiaomei Tuo, Dongli Zhu, Xiaofeng Chen, Yu Rong, Yan Guo, Tielin Yang. The osteoporosis susceptible SNP rs4325274 remotely regulates the SOX6 gene through enhancers [J]. Hereditas(Beijing), 2020, 42(9): 889-897. |
| [10] | Min Chen, Zheng Zhang, Ziyuan Meng, Xuejun Zhang. ATAC-seq and its applications in complex disease [J]. Hereditas(Beijing), 2020, 42(4): 347-353. |
| [11] | Taotao Wang, Yong Yang, Wei Wei, Chentao Lin, Liuyin Ma. Identification and expression analyses of the NAC transcription factor family in Spartina alterniflora [J]. Hereditas(Beijing), 2020, 42(2): 194-211. |
| [12] | Zhaoqing Sun, Bo Yan. The roles and regulation mechanism of transcription factor GATA6 in cardiovascular diseases [J]. Hereditas(Beijing), 2019, 41(5): 375-383. |
| [13] | Haoqiang Yu,Fuai Sun,Wenqi Feng,Fengzhong Lu,Wanchen Li,Fengling Fu. The BES1/BZR1 transcription factors regulate growth, development and stress resistance in plants [J]. Hereditas(Beijing), 2019, 41(3): 206-214. |
| [14] | Junyi Ju,Quan Zhao. Regulation of γ-globin gene expression and its clinical applications [J]. Hereditas(Beijing), 2018, 40(6): 429-444. |
| [15] | Junxia Zou, Ke Chen. Roles and molecular mechanisms of hypoxia-inducible factors in renal cell carcinoma [J]. Hereditas(Beijing), 2018, 40(5): 341-356. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||