遗传 ›› 2022, Vol. 44 ›› Issue (7): 618-628.doi: 10.16288/j.yczz.22-042
• 技术与方法 • 上一篇
李烨荣( ), 吕娟, 王玉国, 谭建新, 邵彬彬, 张菁菁(
), 吕娟, 王玉国, 谭建新, 邵彬彬, 张菁菁( )
)
收稿日期:2022-03-29
修回日期:2022-05-13
出版日期:2022-07-20
发布日期:2022-06-01
通讯作者:
张菁菁
E-mail:lyr15835364548@163.com;18913384682@163.com
作者简介:李烨荣,在读硕士研究生,专业方向:妇产科学,优生学。E-mail: 基金资助:
Yerong Li( ), Juan Lv, Yuguo Wang, Jianxin Tan, Binbin Shao, Jingjing Zhang(
), Juan Lv, Yuguo Wang, Jianxin Tan, Binbin Shao, Jingjing Zhang( )
)
Received:2022-03-29
Revised:2022-05-13
Online:2022-07-20
Published:2022-06-01
Contact:
Zhang Jingjing
E-mail:lyr15835364548@163.com;18913384682@163.com
Supported by:摘要:
脊髓性肌萎缩症(spinal muscular atrophy, SMA)是一种常染色体隐性遗传、儿童致死性神经系统疾病。SMA致病基因为运动神经元存活基因(survival motor neuron1, SMN1)。虽然检测SMN1基因拷贝数的方法众多,但目前适于大规模人群筛查的技术较少。为寻求一种快速准确的实验技术可以用于人群中SMA携带者的大规模筛查,了解区域人群携带情况及常见变异的分布,本研究应用多重竞争性PCR联合毛细管电泳技术检测12例SMA患者及其父母SMN1基因拷贝数,同时对江苏地区151例健康孕妇人群SMN1基因进行拷贝数检测,并通过多重连接依赖探针扩增(multiplex ligation-dependent probe amplification, MLPA)技术验证检测结果。多重竞争性PCR联合毛细管电泳技术结果与MLPA结果一致,显示12例SMA患者均为SMN1基因零拷贝,其父母的SMN1基因拷贝数均为单拷贝,151例健康人群中检测出SMN1基因单拷贝3例(即SMA携带者),占2.0%;SMN1基因双拷贝134例,占88.7%;SMN1基因大于双拷贝14例,占9.3%。因此,多重竞争性PCR联合毛细管电泳技术作为一种快速、简便和准确的检测技术具有着应用于人群中SMA携带者的大规模筛查的潜力。
李烨荣, 吕娟, 王玉国, 谭建新, 邵彬彬, 张菁菁. 应用多重竞争性PCR联合毛细管电泳技术进行脊髓性肌萎缩症携带者筛查[J]. 遗传, 2022, 44(7): 618-628.
Yerong Li, Juan Lv, Yuguo Wang, Jianxin Tan, Binbin Shao, Jingjing Zhang. Application of multiplex competitive PCR combined with capillary electrophoresis in carrier screening of spinal muscular atrophy[J]. Hereditas(Beijing), 2022, 44(7): 618-628.
表1
PCR扩增的基因片段及引物序列"
| 基因片段 | 位置(GRCh37/hg19) | 荧光引物(FAM, 5′→3′) | 反向引物(5′→3′) |
|---|---|---|---|
| 2p | Chr.2:84500611-84500685 | TTGCTTGGAAGGCAGGCAAAC | GTTTCTTTGAGCCAAAAATTCAGAATACAAGGA |
| 20q | Chr.20:35866080-35866163 | TTTTCCCACCAGAGCTTCCCTTAGC | GTTTCATGCAGAACCACCAGGAAAAGG |
| 10p | Chr.10:31120531-31120675 | TTTTTAATGCACACCTCCAGGGAAAAC | GTTTCTTCACTGAGCCCCAGAGACCTGAC |
| 16p | Chr.16:25258133-25258312 | AAAGCTATCGAAAGGTGAGAAATGG | GTTTCCCACCAAGCTGATGTGTTCCTTAG |
| SMN-E2a | Chr.5:69359231-69359324 70234655-70234748 | ACCCTTTCCAGAGCGATGATTCT | GTTTCTTTTTTTGCATTTCATACCTTAAATGAAGCCACA |
| SMN-E1-US | Chr.5:69344093-69344199 70219501-70219607 | TGGTCAACATCATCCCATTCTCC | GTTTCTTAACTCCACGAAAGGAACTTTGAGC |
| SMN-E2b | Chr.5:69361736-69361853 70237160-70237277 | ACTTTTATTCTGTGCACCACCCTGT | GTTTCTTCTTTTAGGTGTGGTTTTTGGTTTACCC |
| SMN-E4 | Chr.5:69363077-69363209 70238501-70238633 | GCAGATTTGGGCTTGATGTTATCTG | GTTTCACACCCTTATAACAAAAACCTGCAT |
| SMN-E6 | Chr.5:69366476-69366612 70241901-70242037 | CCCACCACCTCCCATATGTCC | GTTTCTTAATTGTCAGGAAAAGATGCTGAGTGA |
| SMN-E5 | Chr.5:69365098-69365262 70240521-70240685 | TCTGGCCCAAGGGATGTTCT | GTTTCTTCCACCACCACCACCCCACT |
| NAIP-E4-II | Chr.5:70308186-70308380 | TCCAGATTGTGGGTTCCTTTTGAAC | GTTTCTTAAGCCAGCCTCTGAGAGCACA |
| NAIP-E4-I | Chr.5:70308414-70308621 | AGGAGCAGAAGGAGCGAGCA | GTTTCTTGTGAGGCCGGCACCAAAG |
| NAIP-E5 | Chr.5:70307103-70307328 | GGCCAATGAGATCGGGAGTG | GTTTGTTGGGGAACCATTTGGCATGT |
| SMN-E7-SMN1 | Chr.5:70247563-70247802 | GCAGCCTAATAATTGTTTTCTTTGGG ATAACT | GTTTCTTTTTGTGAGCACCTTCCTTCTTTTTGATTTTGTATG |
| SMN-E8-SMN1 | Chr.5:69374299-69374557 70249719-70249977 | CCACCACACTACCTGGCTGGA | GTTAATGGCAACCCTGCAGAAAAGT |
| SMN-E8-SMN1 | Chr.5:70248466-70248731 | GCCACATACGCCTCACATACATTTTG | GTTTCGGACTCTATTTTGAAAAACCATCTGTAAAAGACAGG |
| SMN-E1-DS-FA | Chr.5:69345517-69345796 70220935-70221214 | ATCAAGGAGCCCAAACTGCTCG | GTTTCTTCGATGAGCAGCGGCGGGAA |
| SMN-E1-DS-FG | Chr.5:69345517-69345796 70220935-70221214 | GTTTCTTTTTCGATGAGCAGCGGCGGAAG | |
| SMN-E7-SMN2 | Chr.5:69372143-69372382 | GCAGCCTAATAATTGTTTTCTTTGGG ATAACT | GTTGTGAGCACCTTCCTTCTTTTTGATTTTGTGTA |
| SMN-E8-SMN2 | Chr.5:69373046-69373311 | GCCACATACGCCTCACATACATTTTG | GTTTCTTTTTTTGGACTCTATTTTGAAAAACCATCTGTAAAAGACGGA |
表2
SMN基因和参考基因的扩增片段信息"
| 反应体系 | 基因片段 | 扩增产物长度(bp) (竞争性DNA/待测样本DNA) | 毛细管电泳参考位置(bp) (竞争性DNA/待测样本DNA) | 扩增片段说明 |
|---|---|---|---|---|
| Pane1 | REF2p | 79/81 | 76.72/78.7 | 参照基因片段 |
| REF10 | 150/152 | 153.96/155.95 | 参照基因片段 | |
| REF16p | 183/185 | 184.32/186.36 | 参照基因片段 | |
| REF20q | 91/93 | 88.76/90.64 | 参照基因片段 | |
| SMN-US | 111/114 | 108.04/111.1 | SMN上游 | |
| SMN-E2a | 103/106 | 100.44/103.29 | SMN外显子2a | |
| SMN-E2b | 122/125 | 118.99/121.84 | SMN外显子2b | |
| SMN-E4 | 133/136 | 131.33/134.22 | SMN外显子4 | |
| SMN-E5 | 169/172 | 169.19/172.3 | SMN外显子5 | |
| SMN-E6 | 141/144 | 140.12/143.58 | SMN外显子6 | |
| SMN1-E7C | 249/252 | 249.91/252.82 | SMN1外显子7(C) | |
| SMN-DS | 159/262 | 260.18/262.99 | SMN下游 | |
| SMN-E8G | 268/271 | 266.62/269.34 | SMN1外显子8(G) | |
| NAIP-E4I | 212/215 | 211.7/214.82 | NAIP外显子4 | |
| NAIP-E4II | 199/202 | 198.5/201.48 | NAIP外显子4 | |
| NAIP-E5 | 229/232 | 229.21/232.29 | NAIP外显子5 | |
| Panel2 | REF2p | 85/87 | 82.33/84.32 | 参照基因片段 |
| REF10p | 157/159 | 160.54/162.5 | 参照基因片段 | |
| REF16p | 191/193 | 189.77/191.89 | 参照基因片段 | |
| SMN2-E7T | 249/252 | 242.83/245.97 | SMN2外显子7(T) | |
| SMN-E8A | 168/171 | 272.88/275.6 | SMN2外显子8(A) |
图3
1例SMA患者及1例SMA携带者的基因型 R-:竞争性DNA片段;A,B:1例SMA患者及标准对照的SMN基因型原始峰图;C,D:1例SMA携带者及标准对照的SMN基因型原始峰图;E:该例SMA患者的SMN基因型柱形图:SMA患者的基因型为SMN1(E7 E8)=0、SMN2(E7 E8)=2、SMN(US E2a E2b E4 E5 E6 DS)=2、NAIP(E4I E4II E5)=1;F:该例SMA携带者的SMN基因型柱形图:SMA携带者的基因型为SMN1(E7 E8)=1、SMN2(E7 E8)=1、SMN(US E2a E2b E4 E5 E6 DS)=2、NAIP(E4I E4II E5)=1。"
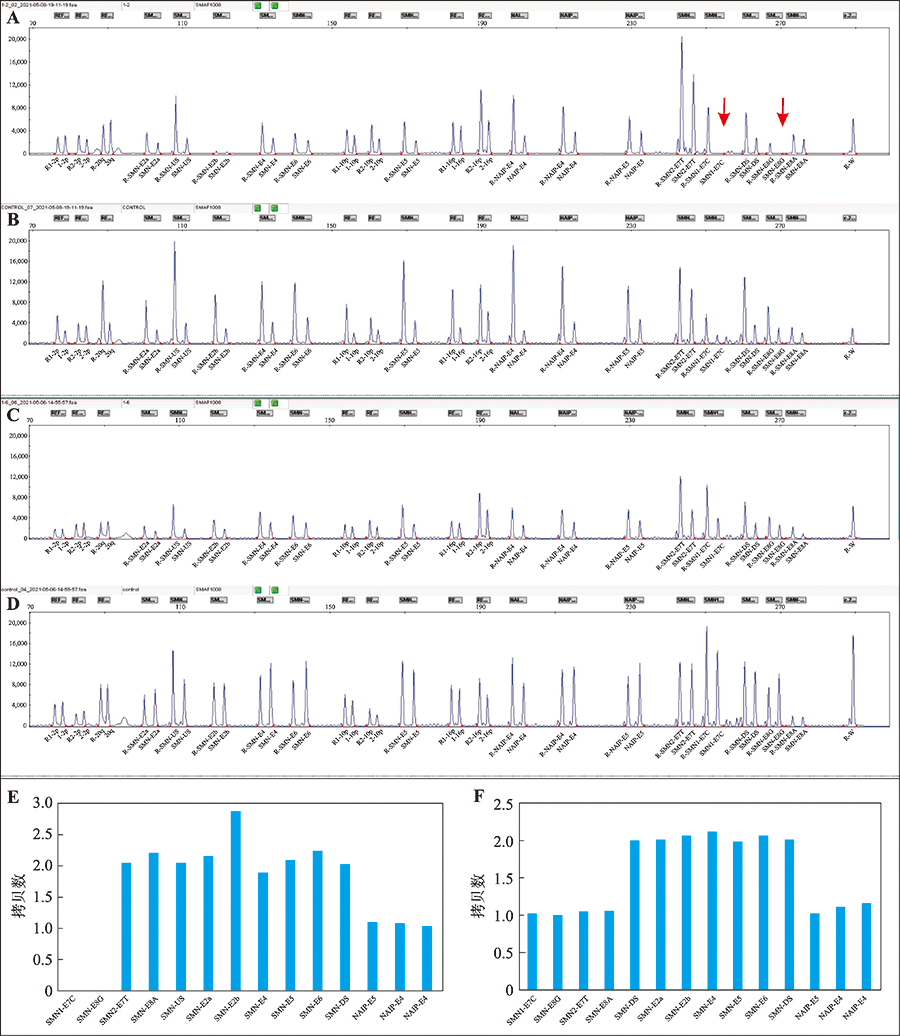
表5
多重竞争性PCR联合毛细管电泳技术、MLPA和荧光定量PCR技术特点对比"
| 特点 | MLPA | 荧光定量PCR技术 | 多重竞争性PCR联合毛细管电泳技术 |
|---|---|---|---|
| 位点 | 对SMN1及SMN2等多重位点进行检测; 含多个参照位点 | 对SMN1单基因检测; 单参照位点 | 对SMN1及SMN2等多重位点进行检测;含3~4个参照位点 |
| 操作性 | 繁琐(操作步骤多,时间久) | 中等 | 简单 |
| 检测周期 | 48 h(耗时、效率低) | 3 h | 4 h |
| 准确性 | 高(CV约10%~20%) DNA质量不一, 波动更大 | 中(CV约10%~20%) 无其他位点辅助验证 | 中(CV约5%)有其他位点辅助验证 |
| 样本要求 | 高(同试剂同批次抽提、检测) | 低 | 低 |
| 结果判读 | 软件判读,需要经验 | 标准化判读 | 计算公式直接得出准确拷贝数 柱形图直观展示 |
| [1] |
Verhaart IEC, Robertson A, Wilson IJ, Aartsma-Rus A, Cameron S, Jones CC, Cook SF, Lochmüller H. Prevalence, incidence and carrier frequency of 5q-linked spinal muscular atrophy-a literature review. Orphanet J Rare Dis, 2017, 12(1):124.
doi: 10.1186/s13023-017-0671-8 pmid: 28676062 |
| [2] |
Bürglen L, Lefebvre S, Clermont O, Burlet P, Viollet L, Cruaud C, Munnich A, Melki J. Structure and organization of the human survival motor neurone (SMN) gene. Genomics, 1996, 32(3):479-482.
pmid: 8838816 |
| [3] |
Qu YJ, Ge XS, Bai JL, Wang LW, Cao YY, Lu YY, Jin YW, Wang H, Song F. Association of copy numbers of survival motor neuron gene 2 and neuronal apoptosis inhibitory protein gene with the natural history in a Chinese spinal muscular atrophy cohort. J Child Neurol, 2015, 30(4):429-436.
doi: 10.1177/0883073814553271 |
| [4] |
Clermont O, Burlet P, Benit P, Chanterau D, Saugier-Veber P, Munnich A, Cusin V. Molecular analysis of SMA patients without homozygous SMN1 deletions using a new strategy for identification of SMN1 subtle mutations. Hum Mutat, 2004, 24(3):417-427.
doi: 10.1002/humu.20092 |
| [5] |
Alías L, Barceló MJ, Bernal S, Martínez-Hernández R, Also-Rallo E, Vázquez C, Santana A, Millán JM, Baiget M, Tizzano EF. Improving detection and genetic counseling in carriers of spinal muscular atrophy with two copies of the SMN1 gene. Clin Genet. 2014, 85(5):470-475.
doi: 10.1111/cge.12222 pmid: 23799925 |
| [6] | Deng KY, Peng JM, Fan HG, Li LL, Xiang YQ. Application of real-time quantitative polymerase chain reaction rapid screening in spinal muscular atrophy smn1 gene carrier. Int J Lab Med, 2016(37):2983-2984. |
| 邓坤仪, 彭建明, 范汉恭, 李丽莲, 向燕群. 荧光定量PCR快速筛查脊髓性肌肉萎缩症smn1致病基因携带者. 国际检验医学杂志, 2016(37):2983-2984. | |
| [7] | Chen LH, Meng YT, Shu JB, Song L. Genetic diagnosis of spinal muscular atrophy using MLPA. Tianjin Med J, 2012, 40(11):1095-1098. |
| 陈红苓, 孟英韬, 舒剑波, 宋力. 运用MLPA进行脊肌萎缩症的基因诊断. 天津医药, 2012, 40(11):1095-1098. | |
| [8] |
Kraszewski JN, Kay DM, Stevens CF, Koval C, Haser B, Ortiz V, Albertorio A, Cohen LL, Jain R, Andrew SP, Young SD, LaMarca NM, Vivo DCD, Caggana M, Chung WK. Pilot study of population-based newborn screening for spinal muscular atrophy in New York state. Genet Med, 2018, 20(6):608-613.
doi: 10.1038/gim.2017.152 pmid: 29758563 |
| [9] |
Hwang HS, Shin GW, Jung GY, Jung GY. A simple and precise diagnostic method for spinal muscular atrophy using a quantitative SNP analysis system. Electrophoresis, 2015, 35(23):3402-3407.
doi: 10.1002/elps.201400207 |
| [10] |
Zhang XQ, Wang B, Zhang LC, You GL, Palais RA, Zhou LM, Fu QH. Accurate diagnosis of spinal muscular atrophy and 22q11.2 deletion syndrome using limited deoxynucleotide triphosphates and high-resolution melting. BMC Genomics, 2018, 19(1):485-492.
doi: 10.1186/s12864-018-4833-4 |
| [11] |
Orlando C, Pinzani P, Pazzagli M. Developments in quantitative PCR. Clin Chem Lab Med, 1998, 36(5):255-269.
pmid: 9676381 |
| [12] |
Hung CC, Lee CN, Lin CY, Cheng WF, Chen CA, Hsieh ST, Yang CC, Jong YJ, Su YN, Lin WL. Identification of deletion and duplication genotypes of the PMP22 gene using PCR-RFLP, competitive multiplex PCR, and multiplex ligation-dependent probe amplification: a comparison. Electrophoresis, 2008, 29(3):618-625.
doi: 10.1002/elps.200700340 |
| [13] |
Brass N, Heckel D, Meese E. Comparative PCR: an improved method to detect gene amplification. Biotechniques, 1998, 24(1):22-24, 26.
pmid: 9454944 |
| [14] |
Wirth B, Herz M, Wetter A, Moskau S, Hahnen E, Rudnik-Schöneborn S, Wienker T, Zerres K. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet, 1999, 64(5):1340-1356.
pmid: 10205265 |
| [15] |
Su YN, Hung CC, Li H, Lee CN, Cheng WF, Tsao PN, Chang MC, Yu CL, Hsieh WS, Lin WL, Hsu SM. Quantitative analysis of SMN1 and SMN2 genes based on DHPLC: a highly efficient and reliable carrier-screening test. Hum Mutat, 2005, 25(5):460-467.
doi: 10.1002/humu.20160 |
| [16] |
Du RQ, Lu CC, Jiang ZW, Li SL, Ma RX, An HJ, Xu MF, An Y, Xia YK, Jin L, Wang XR, Zhang F. Efficient typing of copy number variations in a segmental duplication- mediated rearrangement hotspot using multiplex competitive amplification. J Hum Genet, 2012, 57(8):545-551.
doi: 10.1038/jhg.2012.66 |
| [17] |
Munsat TL, Davies KE. International SMA consortium meeting (26-28 June 1992, Bonn, Germany). Neuromuscul Disord, 1992, 2(5-6):423-428.
doi: 10.1016/S0960-8966(06)80015-5 |
| [18] |
Prior TW, Professional Practice and Guidelines Committee. Carrier screening for spinal muscular atrophy. Genet Med, 2008, 10(11):840-842.
doi: 10.1097/GIM.0b013e318188d069 |
| [19] |
Committee Opinion No. 691: Carrier Screening for Genetic Conditions. Obstet Gynecol, 2017, 129(3):e41-e55.
doi: 10.1097/AOG.0000000000001952 |
| [20] |
Su YN, Hung CC, Lin SY, Chen FY, Chern JPS, Tsai C, Chang TS, Yang CC, Li H, Ho HN, Lee CN. Carrier screening for spinal muscular atrophy (SMA) in 107,611 pregnant women during the period 2005-2009: a prospective population-based cohort study. PLoS One, 2011, 6(2):e17067.
doi: 10.1371/journal.pone.0017067 |
| [21] | Gong B, Zhang L, Hou YP, Hu HY, Li HC, Tan MY, Chen J, Yu J. Carrier screening for spinal muscular atrophy in 4719 pregnant women in Shanghai region. Chin J Med Genet, 2013, 30(6):670-672. |
| 龚波, 章莉, 侯雅萍, 胡荷宇, 李海川, 谭美玉, 陈劲, 俞菁. 中国上海地区4719名孕妇脊髓性肌萎缩症携带者筛查. 中华医学遗传学杂志, 2013, 30(6):670-672. | |
| [22] | Zhang YH, Wang L, He J, Guo JJ, Jin CC, Tang XH, Zhang JM, Chen H, Zhang J, Su J, Zhu BS. Analysis of screening results of spinal muscular atrophy carriers in 3049 people of reproductive age in Yunnan. Chin J Med Genet, 2020, 37(4):384-388. |
| 章印红, 王蕾, 贺静, 郭晶晶, 靳婵婵, 唐新华, 章锦曼, 陈红, 张杰, 苏洁, 朱宝生. 云南地区3049名育龄人群脊髓性肌萎缩症携带者筛查结果分析. 中华医学遗传学杂志, 2020, 37(4):384-388. | |
| [23] | Xu Y, Li Y, Song TT, Zhan Y, Guo FF, Chen BL, Zhang JF. Screening of 6616 carriers of spinal muscular atrophy and analysis of prenatal genetic diagnosis in high-risk fetus. J Pract Obstet Gynecol, 2020, 36(01):47-52. |
| 徐盈, 黎昱, 宋婷婷, 詹瑛, 郭芬芬, 陈必良, 张建芳. 6616例脊髓性肌萎缩症携带者筛查及高风险胎儿产前诊断分析. 实用妇产科杂志, 2020, 36(1):47-52. | |
| [24] | Tan JQ, Zhang X, Wang YL, Luo SQ, Yang FH, Liu BL, Cai R. Screening and prenatal diagnosis of 4931 pregnant women with spinal muscular atrophy mutation carriers in Liuzhou, Guangxi. Chin J Med Genet, 2018, 35(4):467-470. |
| 谭建强, 张旭, 王远流, 罗世强, 杨芳华, 刘百灵, 蔡稔. 广西柳州地区4931例孕妇脊髓性肌萎缩症突变携带者的筛查及产前诊断. 中华医学遗传学杂志, 2018, 35(4):467-470. | |
| [25] |
Huang CH, Chang YY, Chen CH, Kuo YS, Hwu WL, Gerdes T, Ko TM. Copy number analysis of survival motor neuron genes by multiplex ligation-dependent probe amplification. Genet Med. 2007, 9(4):241-248.
doi: 10.1097/GIM.0b013e31803d35bc |
| [26] | Cao YY, Cheng MM, Song F, Qu YJ, Bai JL, Jin YW, Wang H. Familial study of spinal muscular atrophy carriers with SMN1 (2+0) genotype. Hereditas (Beijing), 2021, 43(2):160-168. |
| 曹延延, 程苗苗, 宋昉, 瞿宇晋, 白晋丽, 金煜炜, 王红. 脊髓性肌萎缩症SMN1基因2+0基因型携带者的家系研究. 遗传, 2021, 43(2):160-168. | |
| [27] | Hu HY, Fu XL, Yu J, Zhang B, Tang ZH, Chen HF, Zhang L, Gong B. SMN1 gene deletion analysis using mid-trimester amniotic fluid cells by real-time PCR. Chin J Lab Med, 2016, 39(6):418-422. |
| 胡荷宇, 傅行礼, 俞菁, 张蓓, 唐振华, 陈慧芬, 章莉, 龚波. 利用实时荧光定量PCR分析孕中期羊水细胞SMN1基因缺失. 中华检验医学杂志, 2016, 39(6):418-422. | |
| [28] | Yoon S, Lee CH, Lee KA. Determination of SMN1 and SMN2 copy numbers in a Korean population using multiplex ligation-dependent probe amplification. Korean J Lab Med, 2010, 30(1):93-96. |
| [29] |
Gómez-Curet I, Robinson KG, Funanage VL, Crawford TO, Scavina M, Wang W. Robust quantification of the SMN gene copy number by real-time TaqMan PCR. Neurogenetics, 2007, 8(4):271-278.
pmid: 17647030 |
| [30] |
Zhang JJ, Wang YG, Ma DY, Sun Y, Li YH, Yang PY, Luo CY, Jiang T, Hu P, Xu ZF. Carrier screening and prenatal diagnosis for spinal muscular atrophy in 13,069 Chinese pregnant women. J Mol Diagn, 2020, 22(6):817-822.
doi: 10.1016/j.jmoldx.2020.03.001 |
| [1] | 曹延延, 程苗苗, 宋昉, 瞿宇晋, 白晋丽, 金煜炜, 王红. 脊髓性肌萎缩症SMN1基因 2+0基因型携带者的家系研究[J]. 遗传, 2021, 43(2): 160-168. |
| [2] | 陈万金, 张奇杰, 何瑾, 林翔, 王柠. 脊髓性肌萎缩症患者尿液细胞模型的建立[J]. 遗传, 2014, 36(11): 1168-1172. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
