Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (4): 340-349.doi: 10.16288/j.yczz.20-351
• Research Article • Previous Articles Next Articles
Measuring genetic connectedness between herds based on high density SNP markers
Ziwen Zhou, Xue Wang, Xiangdong Ding( )
)
- National Engineering Laboratory for Animal Breeding, Key Laboratory of Animal Genetics, Breeding and Reproduction of Ministry of Agriculture and Rural Affairs, College of Animal Science and Technology, China Agricultural University,Beijing 100193, China
-
Received:2020-10-19Revised:2021-02-17Online:2021-04-20Published:2021-04-20 -
Contact:Ding Xiangdong E-mail:xding@cau.edu.cn -
Supported by:Supported by China Agriculture Research System No(CARS-35);the National Key Research and Development Project No(2019YFE0106800);Modern Agriculture Science and Technology Key Project of Hebei Province No(19226376D)
Cite this article
Ziwen Zhou, Xue Wang, Xiangdong Ding. Measuring genetic connectedness between herds based on high density SNP markers[J]. Hereditas(Beijing), 2021, 43(4): 340-349.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Summary of genetic connectedness obtained by different approaches based on relationship matrices A,G and H in the 5th generation of simulated data"
| 方法 | 关系矩阵 | ||||
|---|---|---|---|---|---|
| A | G | G0.5 | Gs | H | |
| PEVD | 1.6471 | 1.3218 | 0.9285 | 1.5287 | 1.3725 |
| PEVD(x) | 0.0003 | 0.3248 | 0.2662 | 0.4165 | 0.0016 |
| VED | 0.0019 | 0.3279 | 0.2690 | 0.4196 | 0.0037 |
| CD | 0.5882 | 0.6896 | 0.6790 | 0.7366 | 0.6550 |
| r | 0 | 0.0008 | 0.7478 | NaN | 0.0003 |
| CR | 0 | 0.0031 | 0.9109 | 0.0035 | 0.0200 |
Table 2
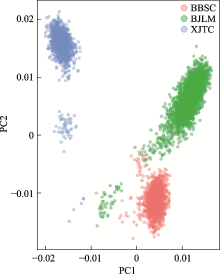
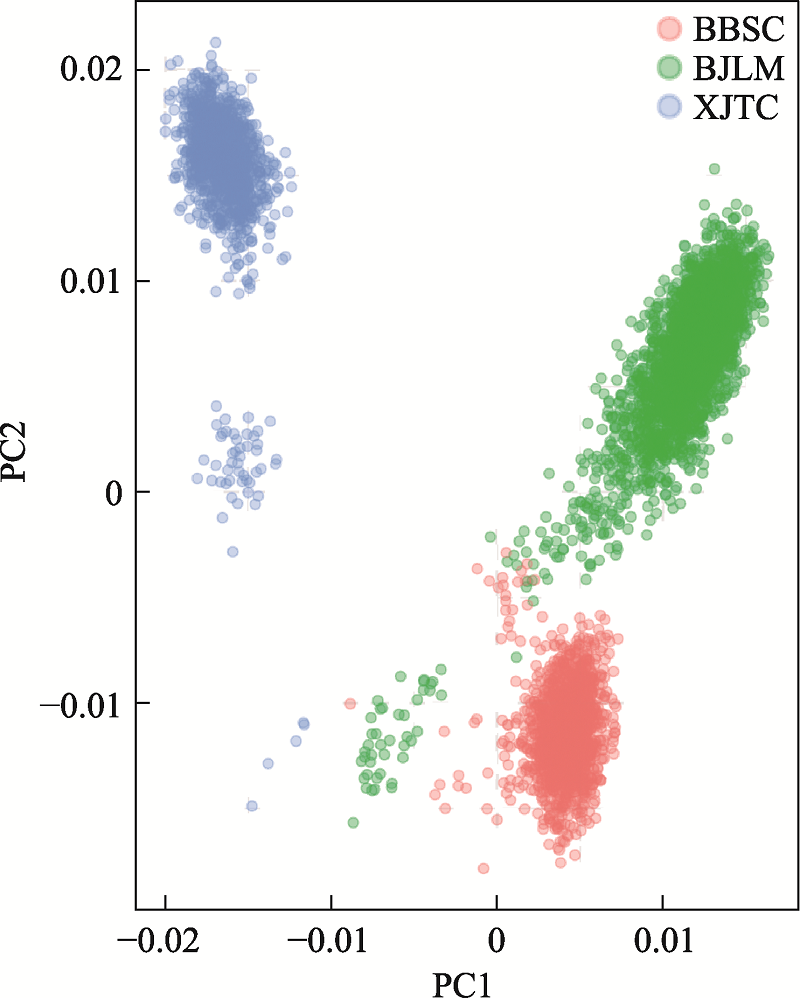
Summary of genetic connectedness obtained by different approaches based on relationship matrices A and G between three pig breeding farms"
| 群体 | 方法 | 关系矩阵 | |||
|---|---|---|---|---|---|
| A | G | Gs | G0.5 | ||
| BJLM-BBSC | PEVD | 15.5600 | 11.9300 | 12.3100 | 9.0500 |
| PEVD(x) | 0.0200 | 1.0600 | 1.1100 | 0.8600 | |
| VED | 0.0500 | 1.0800 | 1.1300 | 0.8800 | |
| CD | 0.5500 | 0.6700 | 0.6800 | 0.6700 | |
| r | 0 | 0.0100 | 0.0200 | 0.5900 | |
| CR | 0 | 0.1500 | 0.1500 | 0.9400 | |
| BJLM-XJTC | PEVD | 15.1500 | 14.1300 | 13.4300 | 11.3200 |
| PEVD(x) | 0.0200 | 2.5200 | 2.3300 | 2.3000 | |
| VED | 0.0400 | 2.5400 | 2.3500 | 2.3300 | |
| CD | 0.5600 | 0.6200 | 0.6100 | 0.6200 | |
| r | 0 | 0.0100 | 0.0100 | 0.4900 | |
| CR | 0 | 0.0700 | 0.0700 | 0.8200 | |
| BBSC-XJTC | PEVD | 13.2300 | 14.5100 | 13.9800 | 12.200 |
| PEVD(x) | 0.0300 | 2.1600 | 2.0500 | 2.4800 | |
| VED | 0.0600 | 2.1900 | 2.0800 | 2.5100 | |
| CD | 0.6200 | 0.6000 | 0.5900 | 0.5900 | |
| r | 0 | 0.0100 | 0.0100 | 0.4800 | |
| CR | 0 | 0.0400 | 0.0400 | 0.8200 | |
"
| 方法 | 关系矩阵 | 遗传力(h2) | |||
|---|---|---|---|---|---|
| 0.1 | 0.3 | 0.5 | 0.7 | ||
| PEVD | A | 1.8900 | 1.6500 | 1.3300 | 0.9200 |
| G | 2.1900 | 1.3200 | 0.8900 | 0.3600 | |
| H | 1.9000 | 1.3700 | 1.0100 | 0.6500 | |
| PEVD(x) | A | 0.0003 | 0.0003 | 0.0003 | 0.0003 |
| G | 0.4000 | 0.3200 | 0.2700 | 0.2200 | |
| H | 0.0095 | 0.0016 | 0.0007 | 0.0003 | |
| VED | A | 0.0063 | 0.0019 | 0.0010 | 0.0006 |
| G | 0.4200 | 0.3300 | 0.2700 | 0.2200 | |
| H | 0.0120 | 0.0040 | 0.0020 | 0.0010 | |
| CD | A | 0.5300 | 0.5900 | 0.6700 | 0.7700 |
| G | 0.4900 | 0.6900 | 0.7900 | 0.9200 | |
| H | 0.5200 | 0.6600 | 0.7500 | 0.8400 | |
| r | A | 0 | 0 | 0 | 0 |
| G | 0.0003 | 0.0008 | NaN | NaN | |
| H | 0.0003 | 0.0003 | NaN | NaN | |
| CR | A | 0 | 0 | 0 | 0 |
| G | 0.0020 | 0.0030 | 0.0030 | 0.0100 | |
| H | 0.1500 | 0.0200 | 0.0090 | 0.0050 | |
| [1] | Mathur PK, Sullivan BP, Chesnais JP. Measuring connectedness: concept and application to a large industry breeding program. Proc 7th World Congr Genet Appl to Livest Prod, 2002,19:23. |
| [2] | Zhang JX, Zhang SY, Qiu XT, Gao H, Wang CC, Wang Y, Zhang Q, Wang ZG, Yang HJ, Ding XD . The genetic connectedness of duroc, landrace and yorkshire pigs in China. Acta Vet Et Zootech Sin, 2017,48(9):1591-1601. |
| 张金鑫, 张锁宇, 邱小田, 高虹, 王长存, 王源, 张勤, 王志刚, 杨红杰, 丁向东 . 我国杜洛克、长白和大白猪场间遗传联系分析. 畜牧兽医学报, 2017,48(9):1591-1601. | |
| [3] | Gao H, Qiu XT, Wang CC, Zhang JX, Zhang SY, Wang Y, Zhang Q, Wang ZG, Yang HJ, Ding XD . The regional joint genetic evaluation of duroc, landrace and yorkshire pigs in China. Acta Vet Et Zootech Sin, 2018,49(12):2567-2575. |
| 高虹, 邱小田, 王长存, 张金鑫, 张锁宇, 王源, 张勤, 王志刚, 杨红杰, 丁向东 . 我国杜洛克、长白、大白猪区域性联合遗传评估研究. 畜牧兽医学报, 2018,49(12):2567-2575. | |
| [4] | Kennedy BW, Trus D. Considerations on genetic connectedness between management units under an animal model. J Anim Sci, 1993,71(9):2341. |
| [5] | Laloë D. Precision and information in linear models of genetic evaluation. Genet Sel Evol, 1993,25(6):557-576. |
| [6] | Lewis RM, Crump RE, Simm G, Thompson R. Assessing connectedness in across-flock genetic evaluations. In: Proceedings of the British Society of Animal Science. Scarborough, 22-24 March, 1999, 121-122. |
| [7] | Mathur PK, Sullivan B, Chesnais J. Estimation of the degree of connectedness between herds or management groups in the canadian swine population. 2002. |
| [8] | Wang AG, Laloe D, Schaeffer LR . Measures of genetic connectedness between herds in swine under mixed linear models. Hereditas (Beijing), 2000,22(5):295-297. |
| 王爱国, Laloe D., Schaeffer LR. 混合线性模型下猪群间遗传联系的度量. 遗传, 2000,22(5):295-297. | |
| [9] |
Muir WM. Comparison of genomic and traditional BLUP-estimated breeding value accuracy and selection response under alternative trait and genomic parameters. J Anim Breed Genet, 2007,124(6):342-355.
pmid: 18076471 |
| [10] |
Daetwyler HD, Villanueva B, Bijma P, Woolliams JA. Inbreeding in genome-wide selection. J Anim Breed Genet, 2007,124(6):369-376.
doi: 10.1111/j.1439-0388.2007.00693.x pmid: 18076474 |
| [11] |
Calus MPL, Meuwissen THE, de Roos APW, Veerkamp RF. Accuracy of genomic selection using different methods to define haplotypes. Genetics, 2008,178(1):553-561.
doi: 10.1534/genetics.107.080838 pmid: 18202394 |
| [12] | Zhang Z, Li X, Ding X, Li J, Zhang Q. GPOPSIM: a simulation tool for whole-genome genetic data. BMC Genet, 2015,16(1):1-6. |
| [13] |
Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet, 2009,84(2):210-223.
doi: 10.1016/j.ajhg.2009.01.005 pmid: 19200528 |
| [14] |
Vanraden PM. Efficient methods to compute genomic predictions. J Dairy Sci, 2008,91(11):4414-4423.
doi: 10.3168/jds.2007-0980 pmid: 18946147 |
| [15] |
Fernando RL, Cheng H, Garrick DJ. An efficient exact method to obtain GBLUP and single-step GBLUP when the genomic relationship matrix is singular. Genet Sel Evol, 2016,48(1):80.
doi: 10.1186/s12711-016-0260-7 pmid: 27788669 |
| [16] | Yu H, Spangler ML, Lewis RM. Genomic relatedness strengthens genetic connectedness across management units. G3-Genes Genom Genet, 2017,7(10):3543-3556. |
| [17] |
Zhang SY, Olasege BS, Liu DY, Wang QS, Pan YC, Ma PP. The genetic connectedness calculated from genomic information and its effect on the accuracy of genomic prediction. PLoS One, 2018,13(7):e0201400.
doi: 10.1371/journal.pone.0201400 pmid: 30063724 |
| [18] | Toro MA, García-Cortés LA, Legarra A. A note on the rationale for estimating genealogical coancestry from molecular markers. Genet Sel Evol, 2011,43(1):1-10. |
| [19] | Vitezica ZG, Aguilar I, Misztal I, Legarra A. Bias in genomic predictions for populations under selection. Genet Res, 2011,93(5):357-366. |
| [20] |
Legarra A, Aguilar I, Misztal I. A relationship matrix including full pedigree and genomic information. J Dairy Sci, 2009,92(9):4656-4663.
pmid: 19700729 |
| [21] | Zhang Q, Ding XD, Chen YS . Development and Application of Swine Genetic Evaluation System in China. Chin J Anim Sci, 2015,51(08):61-65. |
| 张勤, 丁向东, 陈瑶生 . 种猪遗传评估技术研发与评估系统应用. 中国畜牧杂志, 2015,51(08):61-65. | |
| [22] | Mathur PK, Ssllivan BP, Chesnais JP. Measuring connectedness: concept and application to a large industry breeding program. In: Proceedings of 7th world congress on genetics applied to livestock production. Montpellier, 19-23 August, 2002. |
| [23] | Zhang JX, Tang SQ, Song HL, Gao H, Jiang Y, Jiang YF, Mi SR, Meng QL, Yu F, Xiao W, Yun P, Zhang Q, Ding XD . Joint genomic selection of Yorkshire in Beijing. Sci Agric Sin, 2019,52(12):2161-2170. |
| 张金鑫, 唐韶青, 宋海亮, 高虹, 蒋尧, 江一凡, 弥世荣, 孟庆利, 于凡, 肖炜, 云鹏, 张勤, 丁向东 . 北京地区大白猪基因组联合育种研究. 中国农业科学, 2019,52(12):2161-2170. |
| [1] | Zhong Bian, Dongping Cao, Wenshu Zhuang, Shuwei Zhang, Qiaoquan Liu, Lin Zhang. Revelation of rice molecular design breeding: the blend of tradition and modernity [J]. Hereditas(Beijing), 2023, 45(9): 718-740. |
| [2] | Xiaoping Lian, Guangfu Huang, Yujiao Zhang, Jing Zhang, Fengyi Hu, Shilai Zhang. The discovery and utilization of favorable genes in Oryza longistaminata [J]. Hereditas(Beijing), 2023, 45(9): 765-780. |
| [3] | Jun Ma, Anping Fan, Wusheng Wang, Jinchuan Zhang, Xiaojun Jiang, Ruijun Ma, Sheqiang Jia, Fei Liu, Chuchao Lei, Yongzhen Huang. Analysis of genetic diversity and genetic structure of Qinchuan cattle conservation population using whole-genome resequencing [J]. Hereditas(Beijing), 2023, 45(7): 602-616. |
| [4] | Dantong Xu, Yifei Wang, Jiali Cai, Wentao Gong, Xiangchun Pan, Yuhan Tian, Qingpeng Shen, Jiaqi Li, Xiaolong Yuan. Study on detection of CNVs using human whole genome bisulfite sequencing data [J]. Hereditas(Beijing), 2023, 45(4): 324-340. |
| [5] | Zhongsheng Wu, Yu Gao, Yongtao Du, Song Dang, Kangmin He. The protocol of tagging endogenous proteins with fluorescent tags using CRISPR-Cas9 genome editing [J]. Hereditas(Beijing), 2023, 45(2): 165-175. |
| [6] | Meizhen Liu, Liren Wang, Yongmei Li, Xueyun Ma, Honghui Han, Dali Li. Generation of genetically modified rat models via the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2023, 45(1): 78-87. |
| [7] | Xiuli Chen, Haiyan Huang, Qiang Wu. Targeted deletion of 5′HS2 enhancer of β-globin locus control region in K562 cells [J]. Hereditas(Beijing), 2022, 44(9): 783-797. |
| [8] | Mengxuan Xu, Ming Zhou. Advances of RNA polymerase IV in controlling DNA methylation and development in plants [J]. Hereditas(Beijing), 2022, 44(7): 567-580. |
| [9] | Wanjing Ping, Yichen Liu, Qiaomei Fu. Exploring the evolution of archaic humans through sedimentary ancient DNA [J]. Hereditas(Beijing), 2022, 44(5): 362-369. |
| [10] | Jiming Xu, Jianshu Zhu, Mengzhen Li, Han Hu, Chuanzao Mao. Progress on methods for acquiring flanking genomic sequence [J]. Hereditas(Beijing), 2022, 44(4): 313-321. |
| [11] | Cong Liu, Jiani Feng, Weiwei Li, Weiwei Zhu, Yunxin Xue, Dai Wang, Xilin Zhao. Maintenance of dNTP pool homeostasis and genomic stability [J]. Hereditas(Beijing), 2022, 44(2): 96-106. |
| [12] | Cong Zhou, Qiangwei Zhou, Sheng Cheng, Guoliang Li. Research progress of CTCF in mediating 3D genome formation and regulating gene expression [J]. Hereditas(Beijing), 2021, 43(9): 816-821. |
| [13] | Jing Jiang, Anqi Zhao, Tian Xie, Shuwei Chen, Jinsong Li. Construction of genome-wide protein tagging cell and mouse libraries [J]. Hereditas(Beijing), 2021, 43(7): 704-714. |
| [14] | Changgui Lei, Xueyuan Jia, Wenjing Sun. Establish six-gene prognostic model for glioblastoma based on multi-omics data of TCGA database [J]. Hereditas(Beijing), 2021, 43(7): 665-679. |
| [15] | Haitao Wang, Tingting Li, Xun Huang, Runlin Ma, Qiuyue Liu. Application of genetic modification technologies in molecular design breeding of sheep [J]. Hereditas(Beijing), 2021, 43(6): 580-600. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||