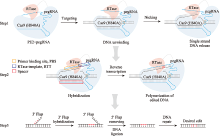
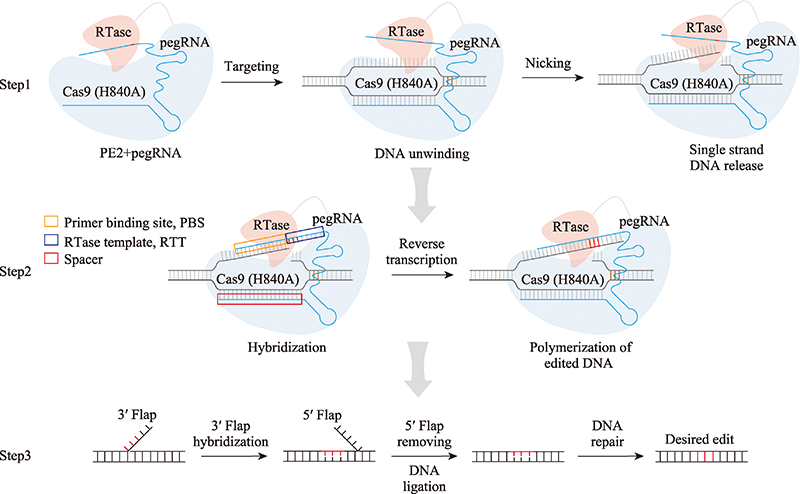
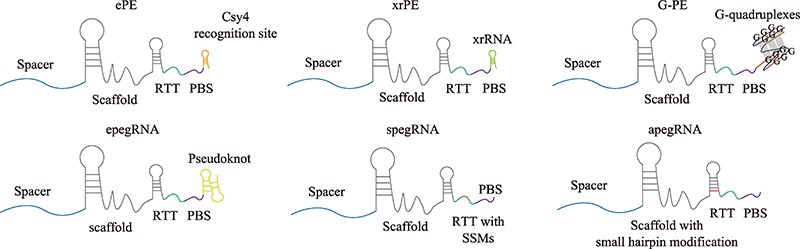
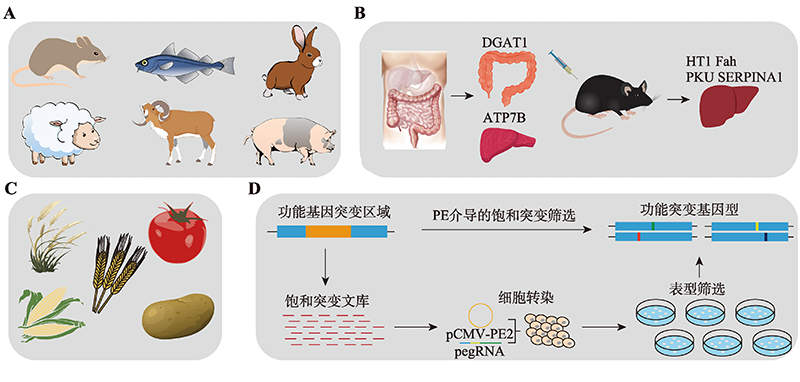
Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (11): 993-1008.doi: 10.16288/j.yczz.22-156
• Review • Previous Articles Next Articles
Prime editing: a search and replace tool with versatile base changes
Yao Liu( ), Xianhui Zhou, Shuhong Huang, Xiaolong Wang(
), Xianhui Zhou, Shuhong Huang, Xiaolong Wang( )
)
- College of Animal Science and Technology/International Joint Research Center of Animal Bio-breeding, Ministry of Agriculture and Rural Affairs/Shaanxi Key Laboratory of Animal Genetic Breeding and Reproduction, Northwest A & F University, Yangling 712100, China
-
Received:2022-07-23Revised:2022-09-01Online:2022-11-20Published:2022-09-16 -
Contact:Wang Xiaolong E-mail:yaoliu@nwafu.edu.cn;xiaolongwang@nwafu.edu.cn -
Supported by:the National Natural Science Foundation of China(31972526);the National Natural Science Foundation of China(32161143010)
Cite this article
Yao Liu, Xianhui Zhou, Shuhong Huang, Xiaolong Wang. Prime editing: a search and replace tool with versatile base changes[J]. Hereditas(Beijing), 2022, 44(11): 993-1008.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Optimized prime editors"
| 版本名称 | 优化策略 | 编辑效率及相应细胞 | 参考文献 |
|---|---|---|---|
| ePE | 抑制pegRNA环化 | >80% (HEK293T) | [ |
| epegRNA | 抑制pegRNA降解 | >80% (HEK293T) | [ |
| xrPE | 抑制pegRNA降解 | >80% (HEK293T) | [ |
| G-PE | 抑制pegRNA降解 | >70% (HEK293T) | [ |
| spegRNA/apegRNA | RTT上引入同义突变/改变pegRNA二级结构 | >80% (HEK293T) | [ |
| — | 增强pegRNA的表达 | >50% (玉米细胞) | [ |
| Dual-pegRNA | 使用两条pegRNA来编码反向互补的逆转录产物 | >40% (水稻细胞) | [ |
| HOPE | 使用成对的pegRNA | >30% (HCT116) | [ |
| CMP-PE3 | 调节染色质结构 | >47% (小鼠胚胎) | [ |
| hyPE2 | 融合表达功能元件 | >43% (HEK293T) | [ |
| PE-P2/PE-P3 | nCas9 N端融合RT,RTT上设计多碱基替换 | >59% (水稻细胞) | [ |
| PEmax | 密码子优化,融合蛋白结构优化等 | >30% (HEK293T) | [ |
| PE2 * | 增加核定位信号 | >6% (小鼠肝脏) | [ |
| ePPE | 删除RT的RNase H结构域,添加病毒核衣壳蛋白 | >25% (水稻细胞) | [ |
| spCas9-NG等 | 利用识别不同PAM的Cas9变体构建PE系统 | >50% (HEK293T) | [ |
| Fn_PE3 | 利用FnCas9构建PE系统 | >80% (HEK293FT) | [ |
| PE4/PE5 | 调节DNA修复途径 | >60% (HEK293T) | [ |
| — | 调节DNA修复途径 | >40% (HEK293) | [ |
Table 2
Technologies derived from prime editing"
| 技术名称 | 改造策略 | 用途 | Cas蛋白 | PAM类型 | 参考文献 |
|---|---|---|---|---|---|
| TwinPE | 两条相向的pegRNA来逆转录出互补的DNA链 | 大片段的替换、插入或删除 | nCas9 | NGG | [ |
| PRIME-Del | 使用一对相向的pegRNA | 精准删除大片段 | nCas9 | NGG | [ |
| GRAND | 使用一对相向的pegRNA | 精准插入长片段 | nCas9 | NGG | [ |
| PEn | Cas9和逆转录酶融合表达 | 小片段插入 | Cas9 | NGG | [ |
| PEDAR | Cas9和逆转录酶融合表达 | 大片段删除与替换 | Cas9 | NGG | [ |
| PEA1 | Cas9和逆转录酶融合,多组分同一质粒表达 | 点突变、插入、删除 | Cas9 | NGG | [ |
| WT-PE | Cas9和逆转录酶融合并结合成对pegRNA | 大片段删除,染色体易位 | Cas9 | NGG | [ |
Table 3
Software for pegRNA designing"
| 软件名称 | 特点 | 网址 | 参考文献 |
|---|---|---|---|
| PrimeDesign | 可进行高通量设计 | http://primedesign.pinellolab.org/ | [ |
| PE-Designer | 设计的同时展示潜在脱靶位点 | http://www.rgenome.net/pe-designer/ | [ |
| Easy-Prime | 基于机器学习优化pegRNA | http://easy-prime.cc/ | [ |
| PlantPegDesigner | 优选双pegRNA策略 | http://www.plantgenomeediting.net | [ |
| pegIT | 设计的同时展示潜在脱靶位点 | https://pegit.giehmlab.dk | [ |
| PINE-CONE | 可进行高通量设计 | https://github.com/xiaowanglab/PINE-CONE | [ |
| PnB Designer | 支持PE、CBE和ABE的gRNA设计 | http://fgcz-shiny.uzh.ch/PnBDesigner/ | [ |
| pegFinder | 可对pegRNA进行打分排序 | http://pegfinder.sidichenlab.org | [ |
| [1] |
Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science, 2013, 339(6121): 819-823.
doi: 10.1126/science.1231143 |
| [2] |
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature, 2016, 533(7603): 420-424.
doi: 10.1038/nature17946 |
| [3] |
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature, 2017, 551(7681): 464-471.
doi: 10.1038/nature24644 |
| [4] |
Ceccaldi R, Rondinelli B, D'Andrea AD. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol, 2016, 26(1): 52-64.
doi: S0962-8924(15)00142-7 pmid: 26437586 |
| [5] | Zong Y, Gao CX. Progress on base editing systems. Hereditas (Beijing), 2019, 41(9): 777-800. |
| 宗媛, 高彩霞. 碱基编辑系统研究进展. 遗传, 2019, 41(9): 777-800. | |
| [6] |
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature, 2019, 576(7785): 149-157.
doi: 10.1038/s41586-019-1711-4 |
| [7] |
Wang XL, Huang XX, Liu Y. Search-and-replace editing of genetic information. Front Agr Sci Eng, 2020, 7(2): 231-232.
doi: 10.15302/J-FASE-2020322 |
| [8] |
Lin QP, Zong Y, Xue CX, Wang SX, Jin S, Zhu ZX, Wang YP, Anzalone AV, Raguram A, Doman JL, Liu DR, Gao CX. Prime genome editing in rice and wheat. Nat Biotechnol, 2020, 38(5): 582-585.
doi: 10.1038/s41587-020-0455-x pmid: 32393904 |
| [9] |
Lin JX, Liu XC, Lu ZY, Huang SS, Wu SS, Yu WX, Liu Y, Zheng XG, Huang XX, Sun Q, Qiao YB, Liu Z. Modeling a cataract disorder in mice with prime editing. Mol Ther Nucleic Acids, 2021, 25 494-501.
doi: 10.1016/j.omtn.2021.06.020 |
| [10] |
Liu Y, Li XY, He ST, Huang SH, Li C, Chen YL, Liu Z, Huang XX, Wang XL. Efficient generation of mouse models with the prime editing system. Cell Discov, 2020, 6(1): 27.
doi: 10.1038/s41421-020-0165-z pmid: 32351707 |
| [11] |
Jang H, Jo DH, Cho CS, Shin JH, Seo JH, Yu G, Gopalappa R, Kim D, Cho SR, Kim JH, Kim HH. Application of prime editing to the correction of mutations and phenotypes in adult mice with liver and eye diseases. Nat Biomed Eng, 2022, 6(2): 181-194.
doi: 10.1038/s41551-021-00788-9 |
| [12] |
Liu Y, Yang G, Huang SH, Li XY, Wang X, Li GL, Chi T, Chen YL, Huang XX, Wang XL. Enhancing prime editing by Csy4-mediated processing of pegRNA. Cell Res, 2021, 31(10): 1134-1136.
doi: 10.1038/s41422-021-00520-x pmid: 34103663 |
| [13] |
Zhang GQ, Liu Y, Huang SS, Qu SY, Cheng DL, Yao Y, Ji QJ, Wang XL, Huang XX, Liu JH. Enhancement of prime editing via xrRNA motif-joined pegRNA. Nat Commun, 2022, 13(1): 1856.
doi: 10.1038/s41467-022-29507-x pmid: 35387980 |
| [14] |
Li XY, Wang X, Sun WJ, Huang SS, Zhong MT, Yao Y, Ji QJ, Huang XX. Enhancing prime editing efficiency by modified pegRNA with RNA G-quadruplexes. J Mol Cell Biol, 2022, 14(4): mjac022.
doi: 10.1093/jmcb/mjac022 |
| [15] |
Nelson JW, Randolph PB, Shen SP, Everette KA, Chen PJ, Anzalone AV, An M, Newby GA, Chen JC, Hsu A, Liu DR. Engineered pegRNAs improve prime editing efficiency. Nat Biotechnol, 2022, 40(3): 402-410.
doi: 10.1038/s41587-021-01039-7 |
| [16] |
Jiang YY, Chai YP, Lu MH, Han XL, Lin QP, Zhang Y, Zhang Q, Zhou Y, Wang XC, Gao CX, Chen QJ. Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol, 2020, 21(1): 257.
doi: 10.1186/s13059-020-02170-5 |
| [17] |
Zhuang Y, Liu JL, Wu H, Zhu QG, Yan YC, Meng HW, Chen PR, Yi CQ. Increasing the efficiency and precision of prime editing with guide RNA pairs. Nat Chem Biol, 2022, 18(1): 29-37.
doi: 10.1038/s41589-021-00889-1 |
| [18] |
Song M, Lim JM, Min S, Oh JS, Kim DY, Woo JS, Nishimasu H, Cho SR, Yoon S, Kim HH. Generation of a more efficient prime editor 2 by addition of the Rad51 DNA-binding domain. Nat Commun, 2021, 12(1): 5617.
doi: 10.1038/s41467-021-25928-2 pmid: 34556671 |
| [19] |
Park SJ, Jeong TY, Shin SK, Yoon DE, Lim SY, Kim SP, Choi J, Lee H, Hong JI, Ahn J, Seong JK, Kim K. Targeted mutagenesis in mouse cells and embryos using an enhanced prime editor. Genome Biol, 2021, 22(1): 170.
doi: 10.1186/s13059-021-02389-w |
| [20] |
Chen PJ, Hussmann JA, Yan J, Knipping F, Ravisankar P, Chen PF, Chen CD, Nelson JW, Newby GA, Sahin M, Osborn MJ, Weissman JS, Adamson B, Liu DR. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell, 2021, 184(22): 5635-5652.e29.
doi: 10.1016/j.cell.2021.09.018 pmid: 34653350 |
| [21] |
Xu W, Yang YX, Yang BY, Krueger CJ, Xiao QL, Zhao S, Zhang L, Kang GT, Wang FP, Yi HM, Ren W, Li L, He XQ, Zhang CM, Zhang B, Zhao JR, Yang JX. A design optimized prime editor with expanded scope and capability in plants. Nat Plants, 2022, 8(1): 455-52.
doi: 10.1038/s41477-022-01143-9 |
| [22] | Zong Y, Liu YJ, Xue CX, Li BS, Li XY, Wang YP, Li J, Liu GW, Huang XX, Cao XF, Gao CX. An engineered prime editor with enhanced editing efficiency in plants. Nat Biotechnol, 2022. |
| [23] | Kweon J, Yoon JK, Jang AH, Shin HR, See JE, Jang GY, Kim JI, Kim Y. Engineered prime editors with PAM flexibility. Mol Ther, 2021, 29(6): 20015-2007. |
| [24] |
Peterka M, Akrap N, Li SY, Wimberger S, Hsieh PP, Degtev D, Bestas B, Barr J, van de Plassche S, Mendoza- Garcia P, Šviković S, Sienski G, Firth M, Maresca M. Harnessing DSB repair to promote efficient homology- dependent and -independent prime editing. Nat Commun, 2022, 13(1): 1240.
doi: 10.1038/s41467-022-28771-1 pmid: 35332138 |
| [25] |
Ferreira da Silva J, Oliveira GP, Arasa-Verge EA, Kagiou C, Moretton A, Timelthaler G, Jiricny J, Loizou JI. Prime editing efficiency and fidelity are enhanced in the absence of mismatch repair. Nat Commun, 2022, 13(1): 760.
doi: 10.1038/s41467-022-28442-1 pmid: 35140211 |
| [26] | Adikusuma F, Lushington C, Arudkumar J, Godahewa GI, Chey YCJ, Gierus L, Piltz S, Geiger A, Jain Y, Reti D, Wilson LOW, Bauer DC, Thomas PQ. Optimized nickase- and nuclease-based prime editing in human and mouse cells. Nucleic Acids Res, 2021, 49(18): 107855-10795. |
| [27] | Tao R, Wang YH, Hu Y, Jiao YG, Zhou LF, Jiang LR, Li L, He XY, Li M, Yu YM, Chen Q, Yao SH. WT-PE: prime editing with nuclease wild-type Cas9 enables versatile large-scale genome editing. Signal Transduct Target Ther, 2022, 7(1): 108. |
| [28] | Wang JL, He Z, Wang GQ, Zhang RW, Duan JY, Gao P, Lei XL, Qiu HY, Zhang CP, Zhang Y, Yin H. Efficient targeted insertion of large DNA fragments without DNA donors. Nat Methods, 2022, 19(3): 3315-340. |
| [29] | Jiang TT, Zhang XO, Weng ZP, Xue W. Deletion and replacement of long genomic sequences using prime editing. Nat Biotechnol, 2022, 40(2): 2275-234. |
| [30] |
Li XS, Zhou LN, Gao BQ, Li GY, Wang X, Wang Y, Wei J, Han WY, Wang ZX, Li JF, Gao RZ, Zhu JJ, Xu WC, Wu J, Yang B, Sun XD, Yang L, Chen J. Highly efficient prime editing by introducing same-sense mutations in pegRNA or stabilizing its structure. Nat Commun, 2022, 13(1): 1669.
doi: 10.1038/s41467-022-29339-9 pmid: 35351879 |
| [31] | Petri K, Zhang WT, Ma JY, Schmidts A, Lee H, Horng JE, Kim DY, Kurt IC, Clement K, Hsu JY, Pinello L, Maus MV, Joung JK, Yeh JJ. CRISPR prime editing with ribonucleoprotein complexes in zebrafish and primary human cells. Nat Biotechnol, 2022, 40(2): 1895-193. |
| [32] | Lin QP, Jin S, Zong Y, Yu H, Zhu ZX, Liu GW, Kou LQ, Wang YP, Qiu JL, Li JY, Gao CX. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat Biotechnol, 2021, 39(8): 9235-927. |
| [33] |
Liu PP, Liang SQ, Zheng CW, Mintzer E, Zhao YG, Ponnienselvan K, Mir A, Sontheimer EJ, Gao GP, Flotte TR, Wolfe SA, Xue W. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat Commun, 2021, 12(1): 2121.
doi: 10.1038/s41467-021-22295-w pmid: 33837189 |
| [34] | Liu B, Dong XL, Cheng HY, Zheng CW, Chen ZX, Rodríguez TC, Liang SQ, Xue W, Sontheimer EJ. A split prime editor with untethered reverse transcriptase and circular RNA template. Nat Biotechnol, 2022. |
| [35] | Zheng CW, Liang SQ, Liu B, Liu PP, Kwan SY, Wolfe SA, Xue W. A flexible split prime editor using truncated reverse transcriptase improves dual-AAV delivery in mouse liver. Mol Ther, 2022, 30(3): 13435-1351. |
| [36] |
Oh Y, Lee WJ, Hur JK, Song WJ, Lee Y, Kim H, Gwon LW, Kim YH, Park YH, Kim CH, Lim KS, Song BS, Huh JW, Kim SU, Jun BH, Jung C, Lee SH. Expansion of the prime editing modality with Cas9 from Francisella novicida. Genome Biol, 2022, 23(1): 92.
doi: 10.1186/s13059-022-02644-8 pmid: 35410288 |
| [37] | Anzalone AV, Gao XD, Podracky CJ, Nelson AT, Koblan LW, Raguram A, Levy JM, Mercer JAM, Liu DR. Programmable deletion, replacement, integration and inversion of large DNA sequences with twin prime editing. Nat Biotechnol, 2022, 40(5): 7315-740. |
| [38] | Choi J, Chen W, Suiter CC, Lee C, Chardon FM, Yang W, Leith A, Daza RM, Martin B, Shendure J. Precise genomic deletions using paired prime editing. Nat Biotechnol, 2022, 40(2): 2185-226. |
| [39] | Bosch JA, Birchak G, Perrimon N. Precise genome engineering in Drosophila using prime editing. Proc Natl Acad Sci USA, 2021, 118(1): e2021996118. |
| [40] |
Qian YQ, Zhao D, Sui TT, Chen M, Liu ZQ, Liu HM, Zhang T, Chen SY, Lai LX, Li ZJ. Efficient and precise generation of Tay-Sachs disease model in rabbit by prime editing system. Cell Discov, 2021, 7(1): 50.
doi: 10.1038/s41421-021-00276-z pmid: 34230459 |
| [41] |
Tong YJ, Jørgensen TS, Whitford CM, Weber T, Lee SY. A versatile genetic engineering toolkit for E. coli based on CRISPR-prime editing. Nat Commun, 2021, 12(1): 5206.
doi: 10.1038/s41467-021-25541-3 |
| [42] | Butt H, Rao GS, Sedeek K, Aman R, Kamel R, Mahfouz M. Engineering herbicide resistance via prime editing in rice. Plant Biotechnol J, 2020, 18(12): 23705-2372. |
| [43] | Geurts MH, de Poel E, Pleguezuelos-Manzano C, Oka R, Carrillo L, Andersson-Rolf A, Boretto M, Brunsveld JE, van Boxtel R, Beekman JM, Clevers H. Evaluating CRISPR-based prime editing for cancer modeling and CFTR repair in organoids. Life Sci Alliance, 2021, 4(10): e202000940. |
| [44] | Li HY, Li JY, Chen JL, Yan L, Xia LQ. Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol Plant, 2020, 13(5): 6715-674. |
| [45] | Lu YM, Tian YF, Shen RD, Yao Q, Zhong DT, Zhang XN, Zhu JK. Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol J, 2021, 19(3): 4155-417. |
| [46] | Wang L, Kaya HB, Zhang N, Rai RT, Willmann MR, Carpenter SCD, Read AC, Martin F, Fei ZJ, Leach JE, Martin GB, Bogdanove AJ.Spelling changes and fluorescent tagging with prime editing vectors for plants. Front Genome Ed, 2021, 3: 617553. |
| [47] |
Perroud PF, Guyon-Debast A, Veillet F, Kermarrec MP, Chauvin L, Chauvin JE, Gallois JL, Nogué F. Prime editing in the model plant physcomitrium patens and its potential in the tetraploid potato. Plant Sci, 2022, 316: 111162.
doi: 10.1016/j.plantsci.2021.111162 |
| [48] | Xu RF, Liu XS, Li J, Qin RY, Wei PC. Identification of herbicide resistance OsACC1 mutations via in planta prime-editing-library screening in rice. Nat Plants, 2021, 7(7): 8885-892. |
| [49] | Erwood S, Bily TMI, Lequyer J, Yan J, Gulati N, Brewer RA, Zhou LC, Pelletier L, Ivakine EA, Cohn RD. Saturation variant interpretation using CRISPR prime editing. Nat Biotechnol, 2022, 40(6): 8855-895. |
| [50] |
Böck D, Rothgangl T, Villiger L, Schmidheini L, Matsushita M, Mathis N, Ioannidi E, Rimann N, Grisch-Chan HM, Kreutzer S, Kontarakis Z, Kopf M, Thöny B, Schwank G. In vivo prime editing of a metabolic liver disease in mice. Sci Transl Med, 2022, 14(636): eabl9238.
doi: 10.1126/scitranslmed.abl9238 |
| [51] | Li JN, Liu Z, Huang SS, Wang X, Li GL, Xu YT, Yu WX, Chen SS, Zhang Y, Ma HH, Ke ZF, Chen J, Sun Q, Huang XX. Efficient base editing in G/C-rich regions to model androgen insensitivity syndrome. Cell Res, 2019, 29(2): 1745-176. |
| [52] |
Liu Z, Lu ZY, Yang G, Huang SS, Li GL, Feng SJ, Liu YJ, Li JN, Yu WX, Zhang Y, Chen J, Sun Q, Huang XX. Efficient generation of mouse models of human diseases via ABE- and BE-mediated base editing. Nat Commun, 2018, 9(1): 2338.
doi: 10.1038/s41467-018-04768-7 pmid: 29904106 |
| [53] |
Gao P, Lyu Q, Ghanam AR, Lazzarotto CR, Newby GA, Zhang W, Choi M, Slivano OJ, Holden K, Walker JA,2nd, Kadina AP, Munroe RJ, Abratte CM, Schimenti JC, Liu DR, Tsai SQ, Long XC, Miano JM. Prime editing in mice reveals the essentiality of a single base in driving tissue-specific gene expression. Genome Biol, 2021, 22(1): 83.
doi: 10.1186/s13059-021-02304-3 pmid: 33722289 |
| [54] | Xiang GH, Ren JL, Hai T, Fu R, Yu DW, Wang J, Li W, Wang HY, Zhou Q. Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs. Cell Mol Life Sci, 2018, 75(24): 46195-4628. |
| [55] | Crispo M, Mulet AP, Tesson L, Barrera N, Cuadro F, dos Santos-Neto PC, Nguyen TH, Crénéguy A, Brusselle L, Anegón I, Menchaca A. Efficient generation of myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes. PLoS One, 2015, 10(8). |
| [56] | Li GW, Zhou SW, Li C, Cai B, Yu HH, Ma BH, Huang Y, Ding YG, Liu Y, Ding Q, He C, Zhou JK, Wang Y, Zhou GX, Li Y, Yan Y, Hua JL, Petersen B, Jiang Y, Sonstegard T, Huang XX, Chen YL, Wang XL. Base pair editing in goat: nonsense codon introgression into FGF5 results in longer hair. FEBS J, 2019, 286(23): 46755-4692. |
| [57] |
Lv QY, Yuan L, Deng JC, Chen M, Wang Y, Zeng J, Li ZJ, Lai LX.Efficient generation of myostatin gene mutated rabbit by CRISPR/Cas9. Sci Rep, 2016, 6: 25029.
doi: 10.1038/srep25029 pmid: 27113799 |
| [58] |
Wang XL, Yu HH, Lei AM, Zhou JK, Zeng WX, Zhu HJ, Dong ZM, Niu YY, Shi BB, Cai B, Liu JW, Huang S, Yan HL, Zhao X, Zhou GX, He XL, Chen XX, Yang YX, Jiang Y, Shi L, Tian X, Wang YJ, Ma BH, Huang XX, Qu L, Chen YL. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci Rep, 2015, 5: 13878.
doi: 10.1038/srep13878 pmid: 26354037 |
| [59] | Whitworth KM, Rowland RRR, Ewen CL, Trible BR, Kerrigan MA, Cino-Ozuna AG, Samuel MS, Lightner JE, McLaren DG, Mileham AJ, Wells KD, Prather RS. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat Biotechnol, 2016, 34(1): 205-22. |
| [60] | Sun JY, Sun KX, Ding YG, Zhou SW, Gao YW, Chen YL, Wang XL. Editing efficiency of four single base editors in sheep (Ovis aries) and goat (Capra hircus) fibroblasts. J Agric Biotechnol, 2021, 29(1): 1695-177. |
| 孙嘉媛, 孙珂欣, 丁一格, 周世卫, 高亚伟, 陈玉林, 王小龙. 四种单碱基编辑器在羊成纤维细胞上的编辑效率. 农业生物技术学报, 2021, 29(1): 1695-177. | |
| [61] | Niu XR, Yin SM, Chen X, Shao TT, Li DL. Gene editing technology and its recent progress in disease therapy. Hereditas (Beijing), 2019, 41(7): 5825-598. |
| 牛煦然, 尹树明, 陈曦, 邵婷婷, 李大力. 基因编辑技术及其在疾病治疗中的研究进展. 遗传, 2019, 41(7): 5825-598. | |
| [62] | Gillmore JD, Gane E, Taubel J, Kao J, Fontana M, Maitland ML, Seitzer J, O'Connell D, Walsh KR, Wood K, Phillips J, Xu YX, Amaral A, Boyd AP, Cehelsky JE, McKee MD, Schiermeier A, Harari O, Murphy A, Kyratsous CA, Zambrowicz B, Soltys R, Gutstein DE, Leonard J, Sepp-Lorenzino L, Lebwohl D. CRISPR-Cas9 in vivo gene editing for transthyretin amyloidosis. N Engl J Med, 2021, 385(6): 4935-502. |
| [63] |
Schene IF, Joore IP, Oka R, Mokry M, van Vugt AHM, van Boxtel R, van der Doef HPJ, van der Laan LJW, Verstegen MMA, van Hasselt PM, Nieuwenhuis EES, Fuchs SA. Prime editing for functional repair in patient- derived disease models. Nat Commun, 2020, 11(1): 5352.
doi: 10.1038/s41467-020-19136-7 pmid: 33097693 |
| [64] | Chemello F, Chai AC, Li H, Rodriguez-Caycedo C, Sanchez-Ortiz E, Atmanli A, Mireault AA, Liu N, Bassel- Duby R, Olson EN. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci Adv, 2021, 7(18). |
| [65] | Kim Y, Hong SA, Yu J, Eom J, Jang K, Yoon S, Hong DH, Seo D, Lee SN, Woo JS, Jeong J, Bae S, Choi D. Adenine base editing and prime editing of chemically derived hepatic progenitors rescue genetic liver disease. Cell Stem Cell, 2021, 28(9): 16145-1624.e5. |
| [66] | Lu YM, Zhu JK. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol Plant, 2017, 10(3): 5235-525. |
| [67] | Zong Y, Wang YP, Li C, Zhang R, Chen KL, Ran YD, Qiu JL, Wang DW, Gao CX. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol, 2017, 35(5): 4385-440. |
| [68] | Hua K, Tao XP, Han PJ, Wang R, Zhu JK. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol Plant, 2019, 12(7): 10035-1014. |
| [69] | Li JY, Sun YW, Du JL, Zhao YD, Xia LQ. Generation of targeted point mutations in rice by a modified CRISPR/ Cas9 system. Mol Plant, 2017, 10(3): 5265-529. |
| [70] | Zhang R, Liu JX, Chai ZZ, Chen S, Bai Y, Zong Y, Chen KL, Li JY, Jiang LJ. Gao CX. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nature Plants, 2019, 5(5): 4805-485. |
| [71] | Qin RY, Wei PC. Prime editing creates a novel dimension of plant precise genome editing. Hereditas (Beijing), 2020, 42(6): 5195-523. |
| 秦瑞英, 魏鹏程. Prime editing引导植物基因组精确编辑新局面. 遗传, 2020, 42(6): 519-523. | |
| [72] | Tang X, Sretenovic S, Ren QR, Jia XY, Li MK, Fan TT, Yin DS, Xiang SY, Guo YC, Liu L, Zheng XL, Qi YP, Zhang Y. Plant prime editors enable precise gene editing in rice cells. Mol Plant, 2020, 13(5): 6675-670. |
| [73] |
Xu RF, Li J, Liu XS, Shan TF, Qin RY, Wei PC. Development of plant prime-editing systems for precise genome editing. Plant Commun, 2020, 1(3): 100043.
doi: 10.1016/j.xplc.2020.100043 |
| [74] | Xu W, Zhang CW, Yang YX, Zhao S, Kang GT, He XQ, Song JL, Yang JX. Versatile nucleotides substitution in plant using an improved prime editing system. Mol Plant, 2020, 13(5): 6755-678. |
| [75] | Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science, 2014, 343(6166): 805-84. |
| [76] | Hanna RE, Hegde M, Fagre CR, DeWeirdt PC, Sangree AK, Szegletes Z, Griffith A, Feeley MN, Sanson KR, Baidi Y, Koblan LW, Liu DR, Neal JT, Doench JG. Massively parallel assessment of human variants with base editor screens. Cell, 2021, 184(4): 10645-1080.e20. |
| [77] | Cuella-Martin R, Hayward SB, Fan X, Chen X, Huang JW, Taglialatela A, Leuzzi G, Zhao JF, Rabadan R, Lu C, Shen Y, Ciccia A. Functional interrogation of DNA damage response variants with base editing screens. Cell, 2021, 184(4): 10815-1097.e19. |
| [78] | Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature, 2014, 513(7516): 1205-123. |
| [79] | Zhi SY, Chen YX, Wu GL, Wen JK, Wu JN, Liu QY, Li Y, Kang R, Hu SH, Wang JH, Liang PP, Huang JJ. Dual-AAV delivering split prime editor system for in vivo genome editing. Mol Ther, 2022, 30(1): 2835-294. |
| [80] | Banskota S, Raguram A, Suh S, Du SW, Davis JR, Choi EH, Wang X, Nielsen SC, Newby GA, Randolph PB, Osborn MJ, Musunuru K, Palczewski K, Liu DR. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell, 2022, 185(2): 2505-265.e16. |
| [81] | Pensado A, Seijo B, Sanchez A. Current strategies for DNA therapy based on lipid nanocarriers. Expert Opin Drug Deliv, 2014, 11(11): 17215-1731. |
| [82] | Lee K, Conboy M, Park HM, Jiang FG, Kim HJ, Dewitt MA, Mackley VA, Chang K, Rao A, Skinner C, Shobha T, Mehdipour M, Liu H, Huang WC, Lan F, Bray NL, Li S, Corn JE, Kataoka K, Doudna JA, Conboy I, Murthy N. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng, 2017, 1: 8895-901. |
| [83] |
Hsu JY, Grünewald J, Szalay R, Shih J, Anzalone AV, Lam KC, Shen MW, Petri K, Liu DR, Joung JK, Pinello L. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Nat Commun, 2021, 12(1): 1034.
doi: 10.1038/s41467-021-21337-7 pmid: 33589617 |
| [84] | Hwang GH, Jeong YK, Habib O, Hong SA, Lim K, Kim JS, Bae S. PE-Designer and PE-Analyzer: web-based design and analysis tools for CRISPR prime editing. Nucleic Acids Res, 2021, 49(W1): W4995-W504. |
| [85] |
Li YC, Chen JJ, Tsai SQ, Cheng Y. Easy-Prime: a machine learning-based prime editor design tool. Genome Biol, 2021, 22(1): 235.
doi: 10.1186/s13059-021-02458-0 pmid: 34412673 |
| [86] | Anderson MV, Haldrup J, Thomsen EA, Wolff JH, Mikkelsen JG. PegIT—a web-based design tool for prime editing. Nucleic Acids Res, 2021, 49(W1): W5055-W509. |
| [87] | Standage-Beier K, Tekel SJ, Brafman DA, Wang X. Prime editing guide RNA eesign automation using PINE- CONE. ACS Synth Biol, 2021, 10(2): 4225-427. |
| [88] |
Siegner SM, Karasu ME, Schroder MS, Kontarakis Z, Corn JE. PnB Designer: a web application to design prime and base editor guide RNAs for animals and plants. BMC Bioinformatics, 2021, 22(1): 101.
doi: 10.1186/s12859-021-04034-6 pmid: 33653259 |
| [89] | Chow RD, Chen JS, Shen J, Chen S. A web tool for the design of prime-editing guide RNAs. Nat Biomed Eng, 2021, 5(2): 1905-194. |
| [90] | Kim HK, Yu G, Park J, Min S, Lee S, Yoon S, Kim HH. Predicting the efficiency of prime editing guide RNAs in human cells. Nat Biotechnol, 2021, 39(2): 1985-206. |
| [91] | Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, Cole MA, Liu DR, Joung JK, Bauer DE, Pinello L. CRISPResso 2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol, 2019, 37(3): 2245-226. |
| [92] | Jin S, Lin QP, Luo YF, Zhu ZX, Liu GW, Li YJ, Chen KL, Qiu JL, Gao CX. Genome-wide specificity of prime editors in plants. Nat Biotechnol, 2021, 39(10): 12925-1299. |
| [93] | Kim DY, Moon SB, Ko JH, Kim YS, Kim D. Unbiased investigation of specificities of prime editing systems in human cells. Nucleic Acids Res, 2020, 48(18): 105765-10589. |
| [94] | Gao RZ, Fu ZC, Li XY, Wang Y, Wei J, Li GY, Wang LJ, Wu J, Huang XX, Yang L, Chen J. Genomic and transcriptomic analyses of prime editing guide RNA-independent off-target effects by prime editors. CRISPR J, 2022, 5(2): 2765-293. |
| [95] | Aida T, Wilde JJ, Yang L, Hou Y, Li M, Xu D, Lin J, Qi P, Lu Z, Feng G. Prime editing primarily induces undesired outcomes in mice. biorxiv, 2020. |
| [96] | Musunuru K, Chadwick AC, Mizoguchi T, Garcia SP, DeNizio JE, Reiss CW, Wang K, Iyer S, Dutta C, Clendaniel V, Amaonye M, Beach A, Berth K, Biswas S, Braun MC, Chen HM, Colace TV, Ganey JD, Gangopadhyay SA, Garrity R, Kasiewicz LN, Lavoie J, Madsen JA, Matsumoto Y, Mazzola AM, Nasrullah YS, Nneji J, Ren H, Sanjeev A, Shay M, Stahley MR, Fan SHY, Tam YK, Gaudelli NM, Ciaramella G, Stolz LE, Malyala P, Cheng CJ, Rajeev KG, Rohde E, Bellinger AM, Kathiresan S. In vivo CRISPR base editing of PCSK 9 durably lowers cholesterol in primates. Nature, 2021, 593(7859): 4295-434. |
| [97] | Koblan LW, Erdos MR, Wilson C, Cabral WA, Levy JM, Xiong ZM, Tavarez UL, Davison LM, Gete YG, Mao XJ, Newby GA, Doherty SP, Narisu N, Sheng QH, Krilow C, Lin CY, Gordon LB, Cao K, Collins FS, Brown JD, Liu DR. In vivo base editing rescues Hutchinson-Gilford progeria syndrome in mice. Nature, 2021, 589(7843): 6085-614. |
| [1] | Bingzheng Wang, Chao Zhang, Jiali Zhang, Jin Sun. Conditional editing of the Drosophila melanogaster genome using single transcripts expressing Cas9 and sgRNA [J]. Hereditas(Beijing), 2023, 45(7): 593-601. |
| [2] | Meizhen Liu, Liren Wang, Yongmei Li, Xueyun Ma, Honghui Han, Dali Li. Generation of genetically modified rat models via the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2023, 45(1): 78-87. |
| [3] | Xiaojun Zhang, Kun Xu, Juncen Shen, Lu Mu, Hongrun Qian, Jieyu Cui, Baoxia Ma, Zhilong Chen, Zhiying Zhang, Zehui Wei. A CRISPR/Cas9-Gal4BD donor adapting system for enhancing homology-directed repair [J]. Hereditas(Beijing), 2022, 44(8): 708-719. |
| [4] | Chong Zhang, Zixuan Wei, Min Wang, Yaosheng Chen, Zuyong He. Editing MC1R in human melanoma cells by CRISPR/Cas9 and functional analysis [J]. Hereditas(Beijing), 2022, 44(7): 581-590. |
| [5] | Ziwen Shi, Qing He, Zhuofan Zhao, Xiaowei Liu, Peng Zhang, Moju Cao. Exploration and utilization of maize male sterility resources [J]. Hereditas(Beijing), 2022, 44(2): 134-152. |
| [6] | Yuting Han, Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni. The cutting edge of gene regulation approaches in model organism Drosophila [J]. Hereditas(Beijing), 2022, 44(1): 3-14. |
| [7] | Guangwu Yang, Yuan Tian. The F-box gene Ppa promotes lipid storage in Drosophila [J]. Hereditas(Beijing), 2021, 43(6): 615-622. |
| [8] | Dingwei Peng, Ruiqiang Li, Wu Zeng, Min Wang, Xuan Shi, Jianhua Zeng, Xiaohong Liu, Yaoshen Chen, Zuyong He. Editing the cystine knot motif of MSTN enhances muscle development of Liang Guang Small Spotted pigs [J]. Hereditas(Beijing), 2021, 43(3): 261-270. |
| [9] | Na Wang, Zhilian Jia, Qiang Wu. RFX5 regulates gene expression of the Pcdhα cluster [J]. Hereditas(Beijing), 2020, 42(8): 760-774. |
| [10] | Guoling Li, Shanxin Yang, Zhenfang Wu, Xianwei Zhang. Recent developments in enhancing the efficiency of CRISPR/Cas9- mediated knock-in in animals [J]. Hereditas(Beijing), 2020, 42(7): 641-656. |
| [11] | Yingnan Chen, Jing Lu. Application of CRISPR/Cas9 mediated gene editing in trees [J]. Hereditas(Beijing), 2020, 42(7): 657-668. |
| [12] | Siyuan Liu, Guoqiang Yi, Zhonglin Tang, Bin Chen. Progress on genome-wide CRISPR/Cas9 screening for functional genes and regulatory elements [J]. Hereditas(Beijing), 2020, 42(5): 435-443. |
| [13] | Liwen Bao, Yiye Zhou, Fanyi Zeng. Advances in gene therapy for β-thalassemia and hemophilia based on the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2020, 42(10): 949-964. |
| [14] | Minting Lin, Lulu Lai, Miao Zhao, Biwei Lin, Xiangping Yao. Construction of a striatum-specific Slc20a2 gene knockout mice model by CRISPR/Cas9 AAV system [J]. Hereditas(Beijing), 2020, 42(10): 1017-1027. |
| [15] | Peifeng Liu, Qiang Wu. Probing 3D genome by CRISPR/Cas9 [J]. Hereditas(Beijing), 2020, 42(1): 18-31. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||