Hereditas(Beijing) ›› 2022, Vol. 44 ›› Issue (11): 1009-1027.doi: 10.16288/j.yczz.22-289
• Review • Previous Articles Next Articles
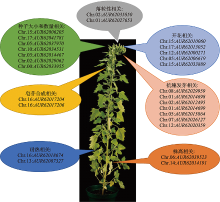
Progress on genomics and locus of important agronomic traits in Chenopodium quinoa
Linghong Li1( ), Tong Gou1, Aixia Ren1, Pengcheng Ding1, Wen Lin1, Xiangyun Wu2, Min Sun1(
), Tong Gou1, Aixia Ren1, Pengcheng Ding1, Wen Lin1, Xiangyun Wu2, Min Sun1( ), Zhiqiang Gao1
), Zhiqiang Gao1
- 1. College of Agronomy, Shanxi Agricultural University, Taigu 030801, Shanxi
2. Shanxi Jiaqi Agri-Tech Co., Ltd., Taiyuan 030006, Shanxi
-
Received:2022-09-02Revised:2022-10-10Online:2022-11-20Published:2022-10-27 -
Contact:Sun Min E-mail:lilinghong00en@163.com;sm_sunmin@126.com -
Supported by:the Fundamental Research Program of Shanxi Province(202103021223158);the Scientific Research Project of Shanxi Province Outstanding Doctoral Work Award Fund(SXBYKY2022022);the Doctoral research Project of Shanxi Agricultural University(2021BQ82)
Cite this article
Linghong Li, Tong Gou, Aixia Ren, Pengcheng Ding, Wen Lin, Xiangyun Wu, Min Sun, Zhiqiang Gao. Progress on genomics and locus of important agronomic traits in Chenopodium quinoa[J]. Hereditas(Beijing), 2022, 44(11): 1009-1027.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 1
Identification and expression analysis of transcription factor gene family in quinoa"
| 名称 | 主要功能 | 成员数量(个) | 组织表达特异性 | 基因功能预测 |
|---|---|---|---|---|
| NAC | 参与调节植物生长发育、次生代谢及胁迫反应 | 107 | 嫩芽、根中特异表达 | AUR62036626、AUR620415254和AUR62043497可能参与藜麦对盐胁迫的响应与适应;AUR62029344可能参与盐胁迫抗性 |
| MADS-box | 参与调控植物花器官发育和开花过程的功能特性 | 103 | 花序、花 | CqMADS33可能具有调控植物开花 |
| WRKY | 参与调控植物应对冻害、干旱、盐害等非生物胁迫与病原菌、虫害等生物胁迫反应 | 92 | 幼苗、茎、叶 | CqWRKY18B-1、CqWRKY21A-1、CqWRKY51A-1和CqWRKY56A-2可能是重要的胁迫调节因子 |
| TH | 参与植物的生长发育和非生物胁迫响应 | 47 | 瘦果、花、叶片中高表达,根茎中低表达 | CqTH36可能参与藜麦的抗逆;CqTH27和CqTH42可能也参与了藜麦花发育的调控 |
| NHX | 参与植物响应盐胁迫的过程 | 46 | NA | 推测35个NHXs基因,参与调控藜麦盐胁迫条件下的离子稳态和盐分分配 |
| TCP | 参与种子萌发、分枝发育、花叶发育、昼夜规律、激素反应及防御反应等过程的调控 | 31 | 叶、花蕾>茎、叶>根、花序、发育中的种子 | TCP11基因在调控叶片发育及开花时间中发挥重要功能;CqTCP18.1基因参与调控分枝发育;CqTCP14在调控叶片发育及干旱胁迫中发挥一定作用;CqTCP15可能在种子萌发过程中发挥一定作用;CqTCP2.1基因可能调控叶片发育 |
| SPL | 参与植物由营养期向花期的过渡、花药发育、芽成熟、开花时期光信号的整合以及维持植物铜稳态等 | 23 | 花序>种子、叶片>茎>幼苗>根 | AUR62011728和AUR62042534可能在干旱胁迫中发挥重要功能 |
| Hsf | 参与植物生长发育,能够响应多种不良逆境 | 23 | 幼苗、茎、叶、花序、干种子 | CqHsf19在干种子中特异表达,可能在种子的发育和成熟过程中起特异的作用 |
| GRF | 参与植物生长、发育、代谢、繁殖、分化等多种生物学过程 | 18 | 种子>花序>根>幼苗>茎>叶片 | AUR62013612和AUR62004236可能参与了藜麦花序的发育过程;AUR62002094和AUR62028212可能与藜麦产量性状有关 |
| WOX | 植物干细胞稳态维持、胚胎发生与胚胎后发育、激素信号转导、初生和次生物质代谢及抗逆响应 | 13 | 茎>种子和叶>幼苗 | AUR62031363、AUR62003747和AUR62035466对氮胁迫有显著响应,可作为氮胁迫响应的候选基因 |
| KEA | 参与植物细胞钾离子的积累和平衡过程 | 10 | 几乎在所有组织中均有表达 | AUR62027069和AUR62030910主要在地上部分发挥功能,可能参与维持植株正常生命活动中的离子稳态和K+利用;AUR62027477、AUR62033999、AUR62023671、AUR62021295、AUR62016085主要在地下部分发挥功能 |
| FAX | 参与质体中脂肪酸向外运输过程 | 10 | 在藜麦不同组织中均存在表达 | AUR62006421、AUR62017596、AUR62026084和AUR62029770在种子中表达水平较高,可能在种子脂肪酸形成和积累中发挥着重要作用 |
| NLP | 参与植物氮素吸收、转运和同化过程 | 9 | NA | AUR62007015、AUR62007012和AUR62019876可能在藜麦氮代谢途径中起重要调控作用 |
Table 2
Summary statistics of genes in quinoa"
| 定位群体 | 性状 | 物理区间(Mb) | 染色体位置 | 候选基因 |
|---|---|---|---|---|
| 310份材料的 全基因组测序 | 茎秆颜色 | 69.72-69.76 | Cq1B | CqCYP76AD1/CqDODA1 |
| 皂苷含量(mg/g) | 8.85-9.20 | Cq5B | AUR62017204/AUR62017206 | |
| 开花日数(天) | 80.50-81.50 | Cq2A | - | |
| 成熟日数(天) | 80.50-81.50 | Cq2A | CqGLX2-2 | |
| 株高(cm) | 80.50-81.50 | Cq2A | - | |
| 穗长(cm) | 80.50-81.50 | Cq2A | - | |
| 千粒重(g) | 63.20-64.87 | Cq8B | CqPP2C/CqRING | |
| 霉变敏感性 | 38.99-39.03 | Cq2A | CqMTA/CqRGA2 |
Table 3
Summary statistics of quantitative trait loci (QTL) in quinoa"
| 定位群体 | 性状 | QTL名称 | 置信区间/遗传位置(cM) | R2(%)-性状 | 置信区间内基因数量 | |
|---|---|---|---|---|---|---|
| PI614889和CHEN-109 组建的F2和F3 | DTF、PH、TKW | pleio4.1 | 69.10-64.44 | 22.01 | 104 | |
| pleio14.1 | 8.95-16.00 | - | 178 | |||
| F3 | PH、PL、PD | pleio20.1 | 10.39-22.00 | 10.86 | 660 | |
| DTF、PH、PD、TKW | pleio4.2 | 69.00-64.59 | 12.44 | 110 | ||
| DTM | dtm3.1 | 35.64-43.00 | 8.73 | 470 | ||
| MS | ms4.1 | 1.68-7.00 | 0.43 | 410 | ||
| ms5.1 | 70.00-76.69 | - | 407 | |||
| PD | pd16.1 | 42.32-44.72 | 20.44 | 47 | ||
| F2 | DTF、DTM、PH, PL、SW、SN、TKW | pleio4.3 | 59.62-62.00 | 16.21 | 88 | |
| DTF、PH、PD、SW | pleio7.1 | 5.32-20.00 | 10.97 | 56 | ||
| SN | sn6.1 | 35.89-37.54 | 5.46 | 313 | ||
| TKW | tkw17.1 | 18.98-23.78 | 8.14 | 156 | ||
| 皂苷 | sap10.1 | 0.00-2.63 | 21.98 | 80 | ||
| sap13.1 | 14.61-20.93 | - | 262 | |||
| sap17.1 | 2.25-10.21 | - | 365 | |||
| Atlas和Red Carina 组建的F3群体 | 籽粒面积 | AR-1 | 23.451 | 11.3 | - | |
| AR-2 | 0 | 9.9 | - | |||
| AR-3 | 4.139 | 9.8 | - | |||
| AR-4 | 81.13 | 8.1 | - | |||
| AR-5 | 27.424 | 7.4 | - | |||
| 花芽出现的时间 | B-1 | 50.867 | 14.7 | - | ||
| B-2 | 0 | 14.4 | - | |||
| B-3 | 48.923 | 11.1 | - | |||
| B-4 | 85.2 | 9.4 | - | |||
| B-5 | 11.475 | 8.3 | - | |||
| B-6 | 80.933 | 7.5 | - | |||
| 色相带1 | C1-1 | 107.778 | 21.6 | - | ||
| C1-2 | 99.488 | 16.2 | - | |||
| C1-3 | 23.425 | 12.3 | - | |||
| C1-4 | 76.2 | 7.9 | - | |||
| C1-5 | 42.401 | 6.6 | - | |||
| C1-6 | 5.491 | 6.5 | - | |||
| C1-7 | 4.048 | 5.4 | - | |||
| 色相带2 蓝光405~470 nm | C2-1 | 105.893 | 18.8 | - | ||
| C2-10 | 36.645 | 4.2 | - | |||
| C2-11 | 2 | 3.5 | - | |||
| C2-2 | 23.425 | 13.9 | - | |||
| C2-3 | 5.491 | 9.9 | - | |||
| C2-4 | 76.2 | 5.9 | - | |||
| Atlas和Red Carina 组建的F3群体 | 色相带2 蓝光405~470 nm | C2-5 | 99.488 | 5.1 | - | |
| C2-6 | 42.401 | 5 | - | |||
| C2-7 | 28.244 | 4.7 | - | |||
| C2-8 | 4.27 | 4.4 | - | |||
| C2-9 | 38.069 | 4.3 | - | |||
| 色相带3 绿光505~590 nm | C3-1 | 105.893 | 19.4 | - | ||
| C3-2 | 110.058 | 7.7 | - | |||
| C3-3 | 71.7 | 7.5 | - | |||
| C3-4 | 99.488 | 6.4 | - | |||
| C3-5 | 42.401 | 6 | - | |||
| C3-6 | 6.643 | 5.5 | - | |||
| C3-7 | 28.884 | 4.8 | - | |||
| 色相带4 红光630~700 nm | C4-1 | 102.512 | 19.5 | - | ||
| C4-2 | 67.9 | 9.3 | - | |||
| C4-3 | 110.058 | 6.8 | - | |||
| C4-4 | 99.488 | 6.2 | - | |||
| C4-5 | 12.732 | 6 | - | |||
| C4-6 | 42.401 | 5.4 | - | |||
| C4-7 | 13.915 | 4.6 | - | |||
| C4-8 | 4.27 | 4.6 | - | |||
| 色相带5 近红外光780 nm | C5-1 | 102.512 | 20.2 | - | ||
| C5-2 | 67.9 | 11.7 | - | |||
| C5-3 | 12.732 | 8.1 | - | |||
| C5-4 | 110.058 | 6 | - | |||
| C5-5 | 52.437 | 5.2 | - | |||
| 色相带6 近红外光 850~890 nm | C6-1 | 102.512 | 11.9 | - | ||
| C6-2 | 52.437 | 11.1 | - | |||
| C6-3 | 12.732 | 9.5 | - | |||
| C6-4 | 116.223 | 8.5 | - | |||
| C6-5 | 67.9 | 7.9 | - | |||
| C6-6 | 11.828 | 7.6 | - | |||
| C6-7 | 110.058 | 7.1 | - | |||
| C6-8 | 91.849 | 5.8 | - | |||
| 色相带7 近红外光 940~970 nm | C7-1 | 11.828 | 14.9 | - | ||
| C7-2 | 51.831 | 12 | - | |||
| C7-3 | 116.223 | 11.4 | - | |||
| C7-4 | 12.732 | 10.6 | - | |||
| C7-5 | 66.9 | 7.4 | - | |||
| C7-6 | 110.058 | 7.3 | - | |||
| C7-7 | 102.512 | 6.4 | - | |||
| C7-8 | 81.526 | 6.3 | - | |||
| Atlas和Red Carina 组建的F3群体 | 混合光系统A | CA-1 | 2.64 | 16.3 | - | |
| CA-2 | 60.805 | 11.2 | - | |||
| CA-3 | 0 | 11.2 | - | |||
| CA-4 | 52.677 | 10.4 | - | |||
| CA-5 | 99.488 | 10.3 | - | |||
| 混合光系统B | CB-1 | 9.892 | 12.5 | - | ||
| CB-2 | 107.778 | 11.5 | - | |||
| CB-3 | 9.784 | 11.1 | - | |||
| CB-4 | 0 | 11 | - | |||
| CB-5 | 6.643 | 9.1 | - | |||
| CB-6 | 29.702 | 8.2 | - | |||
| CB-7 | 3.01 | 7.9 | - | |||
| CB-8 | 7.649 | 7.8 | - | |||
| 混合光系统L | CL-1 | 105.893 | 16.2 | - | ||
| CL-2 | 99.488 | 14.6 | - | |||
| CL-3 | 4.27 | 5.5 | - | |||
| 开花时间 | F9-1 | 19.698 | 14.4 | - | ||
| F9-2 | 110.2 | 13.9 | - | |||
| F9-3 | 3.484 | 11.5 | - | |||
| F9-4 | 16.18 | 9.1 | - | |||
| 产量 | G-1 | 65.4 | 15.5 | - | ||
| G-2 | 25.9 | 13.3 | - | |||
| G-3 | 40.797 | 12.9 | - | |||
| G-4 | 102.57 | 10.8 | - | |||
| G-5 | 7.102 | 10.2 | - | |||
| G-6 | 121.358 | 9.6 | - | |||
| G-7 | 32.724 | 9.2 | - | |||
| G-8 | 56.111 | 6.1 | - | |||
| 种子颜色的色调 | H-1 | 6.643 | 12.3 | - | ||
| H-2 | 107.778 | 9.8 | - | |||
| H-3 | 0 | 8.8 | - | |||
| H-4 | 81.539 | 7.7 | - | |||
| H-5 | 0 | 7.3 | - | |||
| 种子颜色的性状强度 | I-1 | 102.512 | 21.3 | - | ||
| I-10 | 4.27 | 4.4 | - | |||
| I-2 | 23.425 | 15.6 | - | |||
| I-3 | 30.776 | 8.1 | - | |||
| I-4 | 2 | 6.7 | - | |||
| I-5 | 99.488 | 6.3 | - | |||
| I-6 | 50.329 | 5.7 | - | |||
| Atlas和Red Carina 组建的F3群体 | 种子颜色的性状强度 | I-7 | 40.374 | 5.6 | - | |
| I-8 | 8.909 | 5.5 | - | |||
| I-9 | 67.9 | 5.2 | - | |||
| 叶片颜色 | LC-1 | 99.488 | 22.1 | - | ||
| LC-2 | 110.058 | 10.6 | - | |||
| 籽粒长度 | LN-1 | 23.451 | 17.3 | - | ||
| LN-2 | 21.152 | 16.5 | - | |||
| LN-3 | 9.306 | 10.7 | - | |||
| LN-4 | 98.222 | 9.3 | - | |||
| LN-5 | 27.939 | 8.9 | - | |||
| LN-6 | 0 | 8 | - | |||
| 残留菌体 | RB-1 | 105.565 | 15.1 | - | ||
| RB-2 | 32.724 | 15.1 | - | |||
| RB-3 | 40.797 | 11.9 | - | |||
| RB-4 | 34.4 | 10.5 | - | |||
| RB-5 | 60.918 | 9.8 | - | |||
| RB-6 | 29.425 | 9.7 | - | |||
| RB-7 | 24.954 | 8.1 | - | |||
| RB-8 | 102.57 | 7.7 | - | |||
| 性状饱和 | S-1 | 110.006 | 19.4 | - | ||
| S-2 | 99.488 | 7.4 | - | |||
| 千粒重 | T-1 | 3.936 | 19.2 | - | ||
| T-2 | 41.433 | 18.9 | - | |||
| T-3 | 116.108 | 18.6 | - | |||
| T-4 | 27.125 | 13 | - | |||
| T-5 | 44.488 | 10.7 | - | |||
| 籽粒宽度 | W-1 | 81.13 | 13.4 | - | ||
| W-2 | 0 | 11.7 | - | |||
| W-3 | 23.451 | 11.2 | - | |||
| W-4 | 98.222 | 9.8 | - | |||
Table 4
Prediction of genes related to important traits in quinoa"
| 类别 | 性状 | 其他作物中已定位基因 | 藜麦中同源基因 | 所在染色体及物理位置 (Phytozome v1.0) | 序列相似性(%) | 表达部位 |
|---|---|---|---|---|---|---|
| 生长 发育 | 早花 | FT1(Beta vulgaris) | AUR62010060 (CqFT1A) | Chr.15:4930835-4933952 | 81.71 | 花 |
| AUR62013052 (CqFT1B) | Chr.17:79266951-79277600 | 92 | 花 | |||
| FT2 (Beta vulgaris) | AUR62000271 (CqFT2A) | Chr.12:3192361-3196369 | 82.12 | 叶片 | ||
| AUR62006619 (CqFT2B) | Chr.05:77596526-77601590 | 81.56 | 叶片 | |||
| AUR62033889 | Chr.15:31458414-31465667 | 63.79 | NA | |||
| 农艺 性状 | 株高 | Rht-B1(Triticum aestivum)/ RGA1 (Arabidopsis | AUR62039523 | Chr.06:26006908-26013645 | 59.3 | NA |
| Rht-D1(Triticum aestivum) | AUR62014191 | Chr.14:14625033-14626940 | 59.65 | NA | ||
| 种子大小和数量 | GIF1 (Oryza) | AUR62006205 | Chr.15:3135695-3137782 | 60.21 | NA | |
| DA2(Arabidopsis)/ GW2(Oryza) | AUR62041781 | Chr.17:39742130-39752168 | 56.69/45.16 | NA | ||
| AUR62037970 | Chr.05:34646253-34655250 | 56.66/45.57 | NA | |||
| AtCKX5(Arabidopsis)/ Gn1a (Oryza) | AUR62034531 | Chr.10:7564646-7565207 | 68.67/43.49 | NA | ||
| AUR62014467 | Chr.03:4311653-74312220 | 68.67/44.01 | NA | |||
| AtCKX3 (Arabidopsis)/ Gn1a (Oryza) | AUR62029062 | Chr.02:37236856-37237243 | 38.09/43.30 | NA | ||
| AUR62033955 | Chr.00:184848685-184848904 | 35.82/41.65 | NA | |||
| 落粒性 | SHP1/SHP2 (Arabidopsis) | AUR62035850 | Chr.02:11045541-11052900 | 68.64/67.93 | NA | |
| AUR62027653 | Chr.01:128347481-128357581 | 65.68/64.98 | NA | |||
| 品质 | 皂苷合成 | TSARL1 (Medicago truncatula) | AUR62017204 | Chr.16:68549573-68551812 | 32 | 籽粒 |
| TSARL2 (Medicago truncatula) | AUR62017206 | Chr.16:68524854-68527010 | 30.86 | 根 | ||
| 种子贮 藏蛋白 | 11S (A.hypochondriacus) | 11SA | NA | 74 | 籽粒 | |
| 11SB | NA | 74 | 籽粒 | |||
| 非生物 抗性 | 耐盐 | SOS1 | cqSOS1A | NA | NA | 根 |
| cqSOS1B | NA | NA | 根 | |||
| 耐旱 | NA | NA | NA | NA | NA | |
| 耐热 | HSFA1(Arabidopsis) | AUR62018674 | Chr.16:76341712-76354887 | 52.89 | NA | |
| AUR62007327 | Chr.13:2302837-2307436 | 50.87 | NA | |||
| 耐冷 | NA | NA | NA | NA | NA | |
| 抗除草剂 | AHAS | CqAHAS1-CqAHAS6 | NA | NA | NA | |
| 抗穗发芽 | MFT(Arabidopsis) | AUR62029959 | Chr.08:39671124-39679767 | 73.41 | NA | |
| AUR62014698 | Chr.01:29266367-29267601 | 49.13 | NA | |||
| AUR62012495 | Chr.02:4594321-4597301 | 61.21 | NA | |||
| AUR62014699 | Chr.01:29210009-29211182 | 60.47 | NA | |||
| MKK3 (Hordeumvulgare) | AUR62015864 | Chr.05:956636-956737 | 62.03 | NA | ||
| AUR62026127 | Chr.07:82092195-82092329 | 59.96 | NA | |||
| AUR62020359 | Chr.12:56190719-56190853 | 62.55 | NA | |||
| 生物 抗性 | 抗病 | NA | NA | NA | NA | NA |
| 抗虫 | NA | NA | NA | NA | NA |
| [1] | Organization FA. Genebank standards for plant genetic resources for food and agriculture. Genebank Stand Plant Genet Resour Food Agric, 2013, 11(1): 1-16. |
| [2] | Yan F, Li QQ, Dong Y, Ji SD. Industry status and development countermeasures of Chenopodium quinoa. Heilongjiang Agric Sci, 2021, (9): 98-100. |
| 闫锋, 李清泉, 董扬, 季生栋. 藜麦产业现状及发展对策. 黑龙江农业科学, 2021, (9): 98-100. | |
| [3] | Ren GX, Yang XS, Me Y. Current situation of Chinese quinoa industry. Crop Sci, 2015, (5): 1-5. |
| 任贵兴, 杨修仕, 么杨. 中国藜麦产业现状. 作物杂志, 2015, (5): 1-5. | |
| [4] |
Kolano B, McCann J, Orzechowska M, Siwinska D, Temsch E, Weiss-Schneeweiss H. Molecular and cytogenetic evidence for an allotetraploid origin of Chenopodium quinoa and C. berlandieri (Amaranthaceae). Mol Phylogenet Evol, 2016, 100: 109-123.
doi: 10.1016/j.ympev.2016.04.009 |
| [5] |
Yasui Y, Hirakawa H, Oikawa T, Toyoshima M, Matsuzaki C, Ueno M, Mizuno N, Nagatoshi Y, Imamura T, Miyago M, Tanaka K, Mise K, Tanaka T, Mizukoshi H, Mori M, Fujita Y. Draft genome sequence of an inbred line of Chenopodium quinoa, an allotetraploid crop with great environmental adaptability and outstanding nutritional properties. DNA Res, 2016, 23(6): 535-546.
doi: 10.1093/dnares/dsw037 |
| [6] |
Jarvis DE, Ho YS, Lightfoot DJ, Schmöckel SM, Li B, Borm TJA, Ohyanagi H, Mineta K, Michell CT, Saber N, Kharbatia NM, Rupper RR, Sharp AR, Dally N, Boughton BA, Woo YH, Gao G, Schijlen EGWM, Guo XJ, Momin AA, Negrão S, Al-Babili S, Gehring C, Roessner U, Jung C, Murphy K, Arold ST, Gojobori T, van der Linden CG, van Loo EN, Jellen EN, Maughan PJ, Tester M. The genome of Chenopodium quinoa. Nature, 2017, 542(7641): 307-312.
doi: 10.1038/nature21370 |
| [7] | Wang KY, Li L, Li SK, Sun HH, Zhao MZ, Zhang MP, Wang Y. Characterization of the complete chloroplast genome of Chenopodium quinoa Willd. Mitochondrial DNA B Resour, 2017, 2(2): 812-813. |
| [8] |
Stevens MR, Coleman CE, Parkinson SE, Maughan PJ, Zhang HB, Balzotti MR, Kooyman DL, Arumuganathan K, Bonifacio A, Fairbanks DJ, Jellen EN, Stevens JJ. Construction of a quinoa (Chenopodium quinoa Willd.) BAC library and its use in identifying genes encoding seed storage proteins. Theor Appl Genet, 2006, 112(8): 1593-1600.
pmid: 16586115 |
| [9] |
Morales A, Zurita-Silva A, Maldonado J, Silva H. Transcriptional responses of Chilean quinoa (Chenopodium quinoa Willd.) under water deficit conditions uncovers ABA-independent expression patterns. Front Plant Sci, 2017, 8: 216.
doi: 10.3389/fpls.2017.00216 pmid: 28337209 |
| [10] |
Liu Q, Zhang GY, Chen SY. Structure and regulatory function of plant transcription factors. Chin Sci Bull, 2001, 46: 271-278.
doi: 10.1007/BF03187184 |
| [11] | Alshareef NO, Rey E, Khoury H, Tester M, Schmöckel SM. Genome wide identification of NAC transcription factors and their role in abiotic stress tolerance in Chenopodium quinoa. bioRxiv, 2019, Doi: 10.1101/693093. |
| [12] | Zhang DL.Identification and analysis of quinoa MADS-box gene family and study on agrobacterium- mediated root transformation[Dissertation]. Yantai University, 2021. |
| 张东亮.藜麦MADS-box基因家族的鉴定与分析及发根农杆菌介导的根转化研究[学位论文]. 烟台大学, 2021. | |
| [13] |
Yue H, Chang X, Zhi YQ, Wang L, Xing GW, Song WN, Nie XJ. Evolution and identification of the WRKY gene family in quinoa (Chenopodium quinoa). Genes (Basel), 2019, 10(2): 131.
doi: 10.3390/genes10020131 |
| [14] |
Li KY, Fan Y, Zhou GY, Liu XJ, Chen SS, Chang XC, Wu WQ, Duan LL, Yao MX, Wang R, Wang ZL, Yang MF, Ding YQ, Ren MJ, Fan Y, Zhang LY. Genome-wide identification, phylogenetic analysis, and expression profiles of trihelix transcription factor family genes in quinoa (Chenopodium quinoa Willd.) under abiotic stress conditions. BMC Genomics, 2022, 23(1): 499.
doi: 10.1186/s12864-022-08726-y |
| [15] | Zhang YM, Zhu LL, Chen ZG. Identification and expression analysis of NHX gene family in quinoa under salt stress. Biotechnology Bulletin, 2022, 1-10. |
| 张业猛, 朱丽丽, 陈志国. 藜麦NHX基因家族鉴定及盐胁迫下表达分析. 生物技术通报, 2022, 1-10. | |
| [16] | Xiao YL.Identification analysis and functional vertification of TCP gene family in Chenopodium quinoa[Dissertation]. Shandong Normal University, 2021. |
| 肖玉林.藜麦TCP基因家族鉴定分析与功能验证[学位论文]. 山东师范大学, 2021. | |
| [17] | Cao HQ.Isolation and molecular characterization of quinoa SPL family genes[Dissertation]. Shanxi University, 2021. |
| 曹华麒.藜麦SPL家族基因的分离和分子特性分析[学位论文]. 山西大学, 2021. | |
| [18] | Tashi G, Zhan HS, Xing GW, Chang X, Zhang H, Nie XJ, Ji WQ. Genome-wide identification and expression analysis of heat shock transcription factor family in Chenopodium quinoa Willd. Agron, 2018, 8(7): 103. |
| [19] |
Shi PB, He B, Fei YY, Wang J, Wang WY, Wei FY, Lv YD, Gu MF. Identification and expression analysis of GRF transcription factor family of Chenopodium quinoa. Acta Agron Sin, 2019, 45(12): 1841-1850.
doi: 10.3724/SP.J.1006.2019.94049 |
|
时丕彪, 何冰, 费月跃, 王军, 王伟义, 魏福友, 吕远大, 顾闽峰. 藜麦GRF转录因子家族的鉴定及表达分析. 作物学报, 2019, 45(12): 1841-1850.
doi: 10.3724/SP.J.1006.2019.94049 |
|
| [20] | Zhu MX, Yang YS, Yang XL, Zhang F, Yang LY, Wang CY. Identification and expression analysis of WOX transcription factor family of Chenopodium quinoa. J Nat Sci Hunan Norm Univ, 2020, 43(4): 43-49. |
| 朱满喜, 杨雅舒, 杨小兰, 张芳, 杨利艳, 王创云. 藜麦WOX转录因子家族的鉴定及表达分析. 湖南师范大学自然科学学报, 2020, 43(4): 43-49. | |
| [21] | Zhang DL, Wu XL, Tian XQ, Chu J, Guo SL, Chen SH. Identification and expression of CqKEA gene family in Chenopodium quinoa. Journal of Yantai University Natural Science and Engineering Edition, 2021, 34(4): 400-405. |
| 张东亮, 吴筱林, 田晓芹, 褚晶, 郭善利, 陈世华. 藜麦CqKEA基因家族的鉴定及表达. 烟台大学学报(自然科学与工程版), 2021, 34(4): 400-405. | |
| [22] | Shi XD, Sun MH, Wu Q, Tian YS, Xu Y. Identification and expression analysis of fatty acid export gene family in Chenopodium quinoa. Genomics Appl Biol, 2020, 39(12): 5652-5659. |
| 时小东, 孙梦涵, 吴琪, 田银帅, 徐莺. 藜麦脂肪酸转运基因家族FAX的鉴定与表达分析. 基因组学与应用生物学, 2020, 39(12): 5652-5659. | |
| [23] |
Zhu MX, Zhang YR, Yang YS, Yang XL, Wang CY, Deng Y, Zhao L, Zhang LG, Qin LX, Yang LY. Identification and expression analysis of NLP transcription factor family of Chenopodium quinoa Willd. Acta Agric Boreali-Sin, 2021, 36(4): 37-46.
doi: 10.7668/hbnxb.20191913 |
|
朱满喜, 张玉荣, 杨雅舒, 杨小兰, 王创云, 邓妍, 赵丽, 张丽光, 秦丽霞, 杨利艳. 藜麦NLP转录因子家族的鉴定及表达分析. 华北农学报, 2021, 36(4): 37-46.
doi: 10.7668/hbnxb.20191913 |
|
| [24] | Maughan PJ, Smith SM, Rojas-Beltrán JA, Elzinga D, Raney JA, Jellen EN, Bonifacio A, Udall JA, Fairbanks DJ. Single nucleotide polymorphism identification, characterization, and linkage mapping in quinoa. Plant Genome, 2012, 5(3): 114-125. |
| [25] | Fairbanks DJ, Waldrigues A, Ruas CF, Maughan PJ, Robinson LR, Andersen WR, Riede CR, Pauley CS, Caetano LG, Arantes OMN, Fungaro MHP, Vidotto MC, Jankevicius SE. Efficient characterization of biological diversity using field DNA extraction and random amplified polymorphic DNA markers. Braz J Genet, 1993, 16(1): 11-22. |
| [26] |
Maughan PJ, Bonifacio A, Jellen EN, Stevens MR, Coleman CE, Ricks M, Mason SL, Jarvis DE, Gardunia BW, Fairbanks DJ. A genetic linkage map of quinoa (Chenopodium quinoa) based on AFLP, RAPD, and SSR markers. Theor Appl Genet, 2004, 109(6): 1188-1195.
pmid: 15309300 |
| [27] |
Mason SL, Stevens MR, Jellen EN, Bonifacio A, Fairbanks DJ, Coleman CE, Mccarty RR, Rasmussen AG, Maughan PJ. Development and use of microsatellite markers for germplasm characterization in quinoa (Chenopodium quinoa Willd.). Crop Sci, 2005, 45(4): 1618-1630.
doi: 10.2135/cropsci2004.0295 |
| [28] |
Zhang TF, Gu MF, Liu YH, Lv YD, Zhou L, Lu HY, Liang SQ, Bao HB, Zhao H. Development of novel InDel markers and genetic diversity in Chenopodium quinoa through whole-genome re-sequencing. BMC Genomics, 2017, 18(1): 685.
doi: 10.1186/s12864-017-4093-8 pmid: 28870149 |
| [29] |
Jarvis DE, Kopp OR, Jellen EN, Mallory MA, Pattee J, Bonifacio A, Coleman CE, Stevens MR, Fairbanks DJ, Maughan PJ. Simple sequence repeat marker development and genetic mapping in quinoa (Chenopodium quinoa Willd.). J Genet, 2008, 87(1): 39-51.
doi: 10.1007/s12041-008-0006-6 pmid: 18560173 |
| [30] | Cervantes DP, Van Loo E. QTL Mapping for agromorphological traits in quinoa (Chenopodium quinoa Willd.). Wageningen University, 2017. |
| [31] |
Patiranage DSR, Rey E, Emrani N, Wellman G, Schmid K, Schmöckel SM, Tester M, Jung C. Genome-wide association study in quinoa reveals selection pattern typical for crops with a short breeding history. eLife, 2020, 11: e66873.
doi: 10.7554/eLife.66873 |
| [32] |
Maldonado-Taipe N, Barbier F, Schmid K, Jung C, Emrani N. High-density mapping of quantitative trait loci controlling agronomically important traits in quinoa (Chenopodium quinoa Willd.). Front Plant Sci, 2022, 13: 916067.
doi: 10.3389/fpls.2022.916067 |
| [33] |
Pin PA, Nilsson O. The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ, 2012, 35(10): 1742-1755.
doi: 10.1111/j.1365-3040.2012.02558.x |
| [34] |
Golicz AA, Steinfort U, Arya H, Singh MB, Bhalla PL. Analysis of the quinoa genome reveals conservation and divergence of the flowering pathways. Funct Integr Genomics, 2020, 20(2): 245-258.
doi: 10.1007/s10142-019-00711-1 |
| [35] |
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP. 'Green revolution' genes encode mutant gibberellin response modulators. Nature, 1999, 400(6741): 256-261.
doi: 10.1038/22307 |
| [36] | Winkler RG, Freeling M. Physiological genetics of the dominant gibberellin-nonresponsive maize dwarfs, Dwarf8 and Dwarf9. Planta, 1994, 193: 341-348. |
| [37] |
Maliro MFA, Guwela VF, Nyaika J, Murphy KM. Preliminary studies of the performance of quinoa (Chenopodium quinoa Willd.) genotypes under irrigated and rainfed conditions of central Malawi. Front Plant Sci, 2017, 8: 227.
doi: 10.3389/fpls.2017.00227 pmid: 28289421 |
| [38] | Silverstone AL, Ciampaglio CN, Sun T. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell, 1998, 10(2): 155-169. |
| [39] |
Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet, 2007, 39(5): 623-630.
doi: 10.1038/ng2014 |
| [40] |
Wang SG, Ma BT, Gao Q, Jiang GJ, Zhou L, Tu B, Qin P, Tan XQ, Liu PX, Kang YH, Wang YP, Chen WL, Liang CZ, Li SG. Dissecting the genetic basis of heavy panicle hybrid rice uncovered Gn1a and GS3 as key genes. Theor Appl Genet, 2018, 131(6): 1391-1403.
doi: 10.1007/s00122-018-3085-7 |
| [41] |
Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell, 2011, 23(1): 69-80.
doi: 10.1105/tpc.110.079079 |
| [42] |
Ballester P, Ferrándiz C. Shattering fruits: variations on a dehiscent theme. Curr Opin Plant Biol, 2017, 35: 68-75.
doi: S1369-5266(16)30195-9 pmid: 27888713 |
| [43] |
Hofmann NR. SHAT1, a new player in seed shattering of rice. Plant Cell, 2012, 24(3): 839.
doi: 10.1105/tpc.112.240310 |
| [44] |
Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. Shatterproof MADS-box genes control seed dispersal in Arabidopsis. Nature, 2000, 404(6779): 766-770.
doi: 10.1038/35008089 |
| [45] |
Fiallos-Jurado J, Pollier J, Moses T, Arendt P, Barriga- Medina N, Morillo E, Arahana V, de Lourdes Torres M, Goossens A, Leon-Reyes A. Saponin determination, expression analysis and functional characterization of saponin biosynthetic genes in Chenopodium quinoa leaves. Plant Sci, 2016, 250: 188-197.
doi: S0168-9452(16)30094-2 pmid: 27457995 |
| [46] |
Balzotti MRB, Thornton JN, Maughan PJ, Mcclellan DA, Stevens MR, Jellen EN, Fairbanks DJ, Coleman CE. Expression and evolutionary relationships of the Chenopodium quinoa 11S seed storage protein gene. Int J Plant Sci, 2008, 169(2): 281-291.
doi: 10.1086/523874 |
| [47] |
Nakamura S, Ikegami A, Mizuno M, Yagi F, Nomura K. The expression profile of lectin differs from that of seed storage proteins in Castanea crenata trees. Biosci Biotechnol Biochem, 2004, 68(8): 1698-1705.
doi: 10.1271/bbb.68.1698 |
| [48] |
Ruiz KB, Biondi S, Martínez EA, Orsini F, Antognoni F, Jacobsen SE. Quinoa - a model crop for understanding salt-tolerance mechanisms in halophytes. Plant Biosyst, 2016, 150(2): 357-371.
doi: 10.1080/11263504.2015.1027317 |
| [49] | Maughan PJ, Turner TB, Coleman CE, Elzinga DB, Jellen EN, Morales JA, Udall JA, Fairbanks DJ, Bonifacio A. Characterization of salt overly sensitive 1 (SOS1) gene homoeologs in quinoa (Chenopodium quinoa Willd.). Genome, 2009, 52(7): 647-657. |
| [50] |
Schmöckel SM, Lightfoot DJ, Razali R, Tester M, Jarvis DE. Identification of putative transmembrane proteins involved in salinity tolerance in Chenopodium quinoa by integrating physiological data, RNAseq, and SNP analyses. Front Plant Sci, 2017, 8: 1023.
doi: 10.3389/fpls.2017.01023 pmid: 28680429 |
| [51] |
Böhm J, Messerer M, Müller HM, Scholz-Starke J, Gradogna A, Scherzer S, Maierhofer T, Bazihizina N, Zhang H, Stigloher C, Ache P, Al-Rasheid KAS, Mayer KFX, Shabala S, Carpaneto A, Haberer G, Zhu JK, Hedrich R. Understanding the molecular basis of salt sequestration in epidermal bladder cells of Chenopodium quinoa. Curr Biol, 2018, 28(19): 3075-3085.e7.
doi: S0960-9822(18)31049-2 pmid: 30245105 |
| [52] |
Imamura T, Yasui Y, Koga H, Takagi H, Abe A, Nishizawa K, Mizuno N, Ohki S, Mizukoshi H, Mori M. A novel WD40-repeat protein involved in formation of epidermal bladder cells in the halophyte quinoa. Commun Biol, 2020, 3(1): 513.
doi: 10.1038/s42003-020-01249-w pmid: 32943738 |
| [53] |
Liu JX, Wang RM, Liu WY, Zhang HL, Guo YD, Wen RY. Genome-wide characterization of heat-shock protein 70s from Chenopodium quinoa and expression analyses of Cqhsp70s in response to drought stress. Genes (Basel), 2018, 9(2): 35.
doi: 10.3390/genes9020035 |
| [54] |
Golldack D, Li C, Mohan H, Probst N. Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci, 2014, 5: 151.
doi: 10.3389/fpls.2014.00151 pmid: 24795738 |
| [55] |
Liu HC, Liao HT, Charng YY.The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ, 2011, 34(5): 738-751.
doi: 10.1111/j.1365-3040.2011.02278.x |
| [56] |
Hinojosa L, Sanad MNME, Jarvis DE, Steel P, Murphy K, Smertenko A. Impact of heat and drought stress on peroxisome proliferation in quinoa. Plant J, 2019, 99(6): 1144-1158.
doi: 10.1111/tpj.14411 |
| [57] |
Jacobsen SE, Christiansen JL, Rasmussen J. Weed harrowing and inter-row hoeing in organic grown quinoa (Chenopodium quinoa Willd.). Outlook Agric, 2010, 39(3): 223-227.
doi: 10.5367/oa.2010.0001 |
| [58] |
Mestanza U, Riegel R, Silva H, Vásquez SC. Characterization of the acetohydroxyacid synthase multigene family in the tetraploide plant Chenopodium quinoa. Electron J Biotechnol, 2015, 18(6): 393-398.
doi: 10.1016/j.ejbt.2015.07.003 |
| [59] |
Ceccato DV, Bertero HD, Batlla D. Environmental control of dormancy in quinoa (Chenopodium quinoa) seeds: two potential genetic resources for pre-harvest sprouting tolerance. Seed Sci Res, 2011, 21(2): 133-141.
doi: 10.1017/S096025851100002X |
| [60] |
Xi WY, Liu C, Hou XL, Yu H.Mother of FT and TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell, 2010, 22(6): 1733-1748.
doi: 10.1105/tpc.109.073072 |
| [61] |
López-Marqués RL, Nørrevang AF, Ache P, Moog M, Visintainer D, Wendt T, Østerberg JT, Dockter C, Jørgensen ME, Salvador AT, Hedrich R, Gao CX, Jacobsen SE, Shabala S, Palmgren M. Prospects for the accelerated improvement of the resilient crop quinoa. J Exp Bot, 2020, 71(18): 5333-5347.
doi: 10.1093/jxb/eraa285 pmid: 32643753 |
| [62] |
Nakamura S, Pourkheirandish M, Morishige H, Kubo Y, Nakamura M, Ichimura K, Seo S, Kanamori H, Wu JZ, Ando T, Hensel G, Sameri M, Stein N, Sato K, Matsumoto T, Yano M, Komatsuda T. Mitogen-activated protein kinase kinase 3 regulates seed dormancy in barley. Curr Biol, 2016, 26(6): 775-781.
doi: 10.1016/j.cub.2016.01.024 pmid: 26948880 |
| [63] |
Danielsen S, Bonifacio A, Ames T. Diseases of quinoa (Chenopodium quinoa). Food Rev Int, 2006, 19(1-2): 43-59.
doi: 10.1081/FRI-120018867 |
| [64] | Khalifa W, Thabet M. Variation in downy mildew (Peronospora variabilis Gäum) resistance of some quinoa (Chenopodium quinoa Willd.) cultivars under Egyptian conditions. Middle East J Agric, 2018, 7(2): 671-682. |
| [65] | Mhada M, Ezzahiri B, Benlhabib O. Assessment of downy mildew resistance (Peronospora farinosa) in a quinoa (Chenopodium quinoa Willd.) germplasm. Int J Agric Biol Eng, 2014, 8(3): 277-280. |
| [66] |
Ochoa J, Frinking HD, Jacobs T. Postulation of virulence groups and resistance factors in the quinoa/downy mildew pathosystem using material from Ecuador. Plant Pathol, 1999, 48(3): 425-430.
doi: 10.1046/j.1365-3059.1999.00352.x |
| [67] |
Rasmussen C, Lagnaoui A, Esbjerg P. Advances in the knowledge of quinoa pests. Food Rev Int, 2003, 19(1-2): 61-75.
doi: 10.1081/FRI-120018868 |
| [68] |
Pereira E, Encina-Zelada C, Barros L, Gonzales-Barron U, Cadavez V, Ferreira ICFR. Chemical and nutritional characterization of quinoa (Chenopodium quinoa Willd.) grains: A good alternative to nutritious food. Food Chem, 2019, 280: 110-114.
doi: S0308-8146(18)32172-1 pmid: 30642475 |
| [69] |
Ruiz KB, Biondi S, Oses R, Acuña-Rodríguez IS, Antognoni F, Martinez-Mosqueira EA, Coulibaly A, Canahua-Murillo A, Pinto M, Zurita-Silva A, Bazile D, Jacobsen SE, Molina-Montenegro MA. Quinoa biodiversity and sustainability for food security under climate change. A review. Agron Sustainable Dev, 2014, 34(2): 349-359.
doi: 10.1007/s13593-013-0195-0 |
| [70] |
Zurita-Silva A, Fuentes F, Zamora P, Jacobsen SE, Schwember AR. Breeding quinoa (Chenopodium quinoa Willd.): potential and perspectives. Mol Breed, 2014, 34(1): 13-30.
doi: 10.1007/s11032-014-0023-5 |
| [71] | Lin C, Liu ZJ, Dong YM, Michel V, Mao ZC. Domesticated cultivation and genetic breeding of Chenopodium quinoa. Hereditas(Beijing), 2019, 41(11): 1009-1022. |
| 林春, 刘正杰, 董玉梅, Michel Vales, 毛自朝. 藜麦的驯化栽培与遗传育种. 遗传, 2019, 41(11): 1009-1022. | |
| [72] |
Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, Wang B, Yang ZF, Li HY, Lin YR, Xie YY, Shen RX, Chen SF, Wang Z, Chen YL, Guo JX, Chen LT, Zhao XC, Dong ZC, Liu YG. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in Monocot and Dicot plants. Mol Plant, 2015, 8(8): 1274-1284.
doi: 10.1016/j.molp.2015.04.007 pmid: 25917172 |
| [1] | Qianwen Lv, Yongfang Yang. The biological functions of peptide signaling in plant and the advances on its utilization for crop improvement [J]. Hereditas(Beijing), 2023, 45(9): 813-828. |
| [2] | Xiaoping Lian, Guangfu Huang, Yujiao Zhang, Jing Zhang, Fengyi Hu, Shilai Zhang. The discovery and utilization of favorable genes in Oryza longistaminata [J]. Hereditas(Beijing), 2023, 45(9): 765-780. |
| [3] | Wenrui Shi, Hongzhu Qu, Xiangdong Fang. Overview of multi-omics research in gout [J]. Hereditas(Beijing), 2023, 45(8): 643-657. |
| [4] | Yi Zhang, Zhi-Ying Wu. Pathogenesis and therapeutic advances of cerebral autosomal- dominant arteriopathy with subcortical infarcts and leukoencephalopathy [J]. Hereditas(Beijing), 2023, 45(7): 568-579. |
| [5] | Chenghao Yan, Weiyu Bai, Zhimeng Zhang, Junling Shen, Youjun Wang, Jianwei Sun. The roles and mechanism of STIM1 in tumorigenesis and metastasis [J]. Hereditas(Beijing), 2023, 45(5): 395-408. |
| [6] | Shunze Wang, Feng Jiang, Dongli Zhu, Tie-Lin Yang, Yan Guo. Application of Hi-C technology in three-dimensional genomics research and disease pathogenesis analysis [J]. Hereditas(Beijing), 2023, 45(4): 279-294. |
| [7] | Wenlong Wang, Chunxia Zhang. Research progress on the study of transcriptome-wide poly(A) tails in mammalian oocytes and early embryos [J]. Hereditas(Beijing), 2023, 45(4): 273-278. |
| [8] | Dong Chang, Xiangxiang Liu, Rui Liu, Jianwei Sun. The role and regulatory mechanism of FSCN1 in breast tumorigenesis and progression [J]. Hereditas(Beijing), 2023, 45(2): 115-127. |
| [9] | Biyuan Li, Yanting Zhao, Zhichen Yue, Juanli Lei, Qizan Hu, Peng Tao. Identification of DELLA gene family in head cabbage and analysis of mRNA transport in the heterograft [J]. Hereditas(Beijing), 2023, 45(2): 156-164. |
| [10] | Dandan Wu, Mingkun Zhu, Zhongyan Fang, Wei Ma. Progress on molecular composition and genetic mechanism of plant B chromosomes [J]. Hereditas(Beijing), 2022, 44(9): 772-782. |
| [11] | Mingliang Jiang, Hong Lang, Xiaonan Li, Ye Zu, Jing Zhao, Shenling Peng, Zhen Liu, Zongxiang Zhan, Zhongyun Piao. Progress on plant orphan genes [J]. Hereditas(Beijing), 2022, 44(8): 682-694. |
| [12] | Chong Zhang, Zixuan Wei, Min Wang, Yaosheng Chen, Zuyong He. Editing MC1R in human melanoma cells by CRISPR/Cas9 and functional analysis [J]. Hereditas(Beijing), 2022, 44(7): 581-590. |
| [13] | Yan Guo, Lele Yang, Huayu Qi. Transcriptome analysis of mouse male germline stem cells reveals characteristics of mature spermatogonial stem cells [J]. Hereditas(Beijing), 2022, 44(7): 591-608. |
| [14] | Zichuan Wang, Jiaqi Zhang, Lei Li. In vitro investigation of mammalian early embryonic development [J]. Hereditas(Beijing), 2022, 44(4): 269-274. |
| [15] | Tian Shu, Haochang Hu, Caijie Shen, Shaoyi Lin, Xiaomin Chen. Research progress of the correlation between genotype and phenotype in hypertrophic cardiomyopathy [J]. Hereditas(Beijing), 2022, 44(3): 198-207. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||