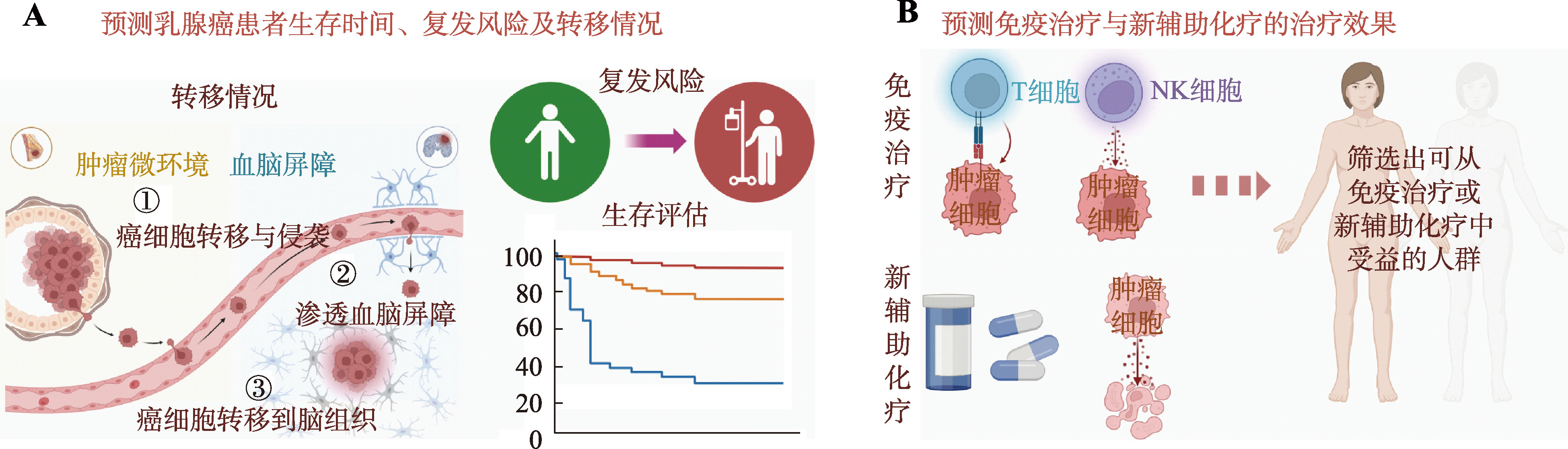
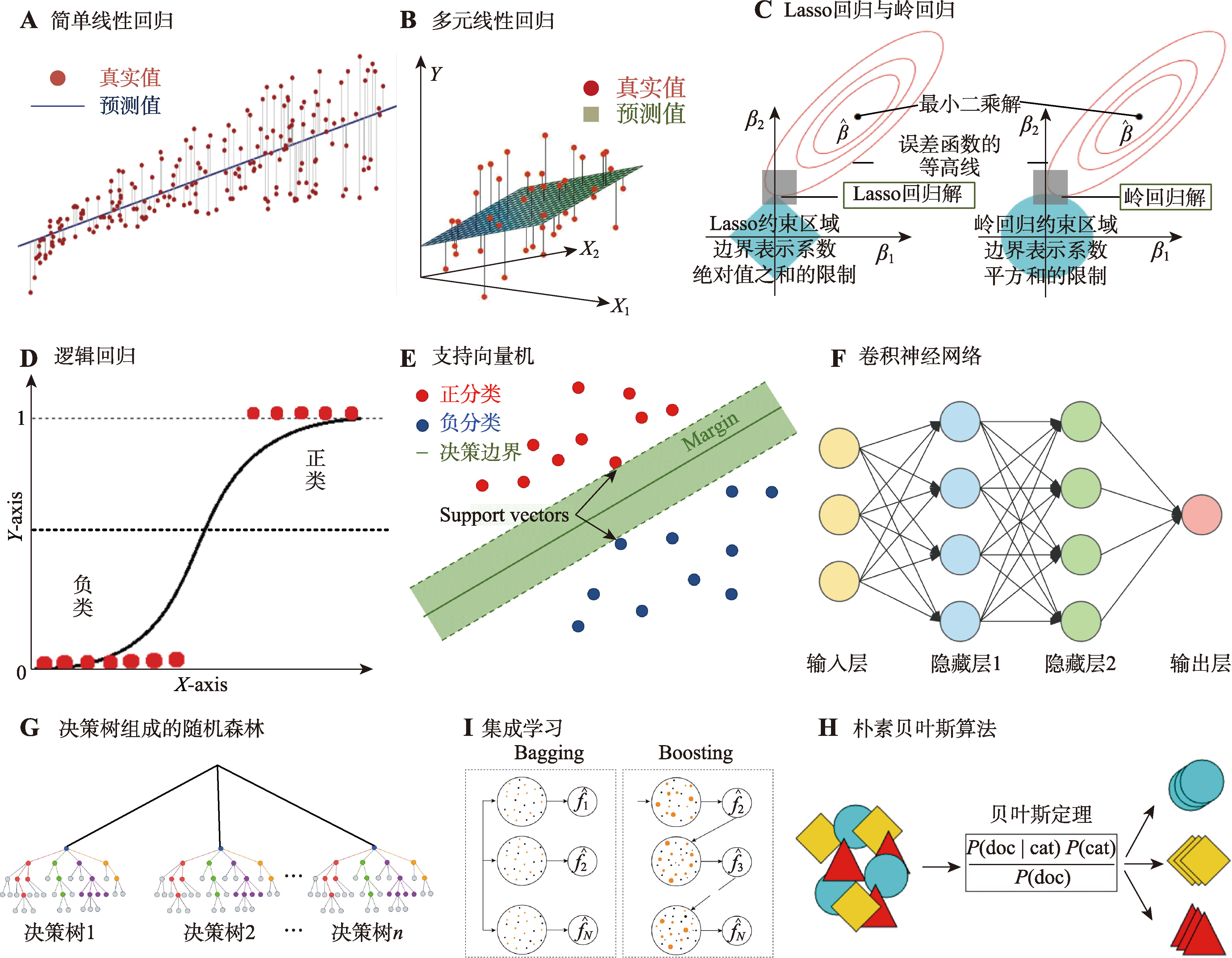
Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (10): 820-832.doi: 10.16288/j.yczz.24-156
• Review • Previous Articles Next Articles
Machine learning applications in breast cancer survival and therapeutic outcome prediction based on multi-omic analysis
Ziyi Zhang1,2( ), Qilin Wang1,2, Junyou Zhang1,2, Yingying Duan1,2, Jiaxin Liu1,2, Zhaoshuo Liu1,2, Chunyan Li1,2,3,4(
), Qilin Wang1,2, Junyou Zhang1,2, Yingying Duan1,2, Jiaxin Liu1,2, Zhaoshuo Liu1,2, Chunyan Li1,2,3,4( )
)
- 1. School of Engineering Medicine, Beihang University, Beijing 100191, China
2. School of Biological Science and Medical Engineering, Beihang University, Beijing 100191, China
3. Key Laboratory of Big Data-Based Precision Medicine (Ministry of Industry and Information Technology), Beihang University, Beijing 100191, China
4. Beijing Advanced Innovation Center for Big Data-Based Precision Medicine, Beihang University, Beijing 100191, China
-
Received:2024-05-31Revised:2024-08-18Online:2024-08-19Published:2024-08-19 -
Contact:Chunyan Li E-mail:zhangziyi@buaa.edu.cn;lichunyan@buaa.edu.cn -
Supported by:National Natural Science Foundation of China(32270610);National Natural Science Foundation of China(82072499);National Natural Science Foundation of China(31801094);Fundamental Research Funds for the Central Universities(YWF-21-BJ-J-T105)
Cite this article
Ziyi Zhang, Qilin Wang, Junyou Zhang, Yingying Duan, Jiaxin Liu, Zhaoshuo Liu, Chunyan Li. Machine learning applications in breast cancer survival and therapeutic outcome prediction based on multi-omic analysis[J]. Hereditas(Beijing), 2024, 46(10): 820-832.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 2
The brief summary of multi-omics data"
| 分型 | 研究对象 |
|---|---|
| 基因组学(genomics) | 所有遗传物质的结构功能与遗传变异 |
| 转录组学(transcriptomics) | 特定条件下DNA转录为RNA的基因表达调控 |
| 表观组学(epigenomics) | 包含DNA甲基化、组蛋白修饰在内的基因表达调控的表观遗传机制 |
| 蛋白组学(proteomics) | 所有蛋白质的组成、结构和功能 |
| 代谢组学(metabolomics) | 代谢产物的组成与代谢通路的调控 |
| 免疫组学(immunomics) | 应对疾病时免疫系统调节的机制 |
| 病理组学(pathomics) | 疾病的形态学特征及组织和细胞的结构与功能 |
| [1] | Han BF, Zheng RS, Zeng HM, Wang SM, Sun KX, Chen R, Li L, Wei WQ, He J. Cancer incidence and mortality in China, 2022. J Natl Cancer Cent, 2024, 4(1): 47-53. |
| [2] |
Hapach LA, Carey SP, Schwager SC, Taufalele PV, Wang WJ, Mosier JA, Ortiz-Otero N, McArdle TJ, Goldblatt ZE, Lampi MC, Bordeleau F, Marshall JR, Richardson IM, Li JH, King MR, Reinhart-King CA. Phenotypic heterogeneity and metastasis of breast cancer cells. Cancer Res, 2021, 81(13): 3649-3663.
doi: 10.1158/0008-5472.CAN-20-1799 pmid: 33975882 |
| [3] |
Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res, 2020, 22(1): 61.
doi: 10.1186/s13058-020-01296-5 pmid: 32517735 |
| [4] |
Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biol Res, 2017, 50(1): 33.
doi: 10.1186/s40659-017-0140-9 pmid: 28969709 |
| [5] | Kundu M, Butti R, Panda VK, Malhotra D, Das S, Mitra T, Kapse P, Gosavi SW, Kundu GC. Modulation of the tumor microenvironment and mechanism of immunotherapy- based drug resistance in breast cancer. Mol Cancer, 2024, 23(1): 92. |
| [6] | Rakha EA, Green AR. Molecular classification of breast cancer: what the pathologist needs to know. Pathology, 2017, 49(2): 111-119. |
| [7] |
Huang SG, Yang J, Shen N, Xu QS, Zhao Q. Artificial intelligence in lung cancer diagnosis and prognosis: Current application and future perspective. Semin Cancer Biol, 2023, 89: 30-37.
doi: 10.1016/j.semcancer.2023.01.006 pmid: 36682439 |
| [8] |
Jin X, Zhou YF, Ma D, Zhao S, Lin CJ, Xiao Y, Fu T, Liu CL, Chen YY, Xiao WX, Liu YQ, Chen QW, Yu Y, Shi LM, Shi JX, Huang W, Robertson JFR, Jiang YZ, Shao ZM. Molecular classification of hormone receptor-positive HER2-negative breast cancer. Nat Genet, 2023, 55(10): 1696-1708.
doi: 10.1038/s41588-023-01507-7 pmid: 37770634 |
| [9] |
Li RQ, Yan L, Zhang L, Ma HX, Wang HW, Bu P, Xi YF, Lian J. Genomic characterization reveals distinct mutational landscapes and therapeutic implications between different molecular subtypes of triple-negative breast cancer. Sci Rep, 2024, 14(1): 12386.
doi: 10.1038/s41598-024-62991-3 pmid: 38811720 |
| [10] |
Spring LM, Fell G, Arfe A, Sharma C, Greenup R, Reynolds KL, Smith BL, Alexander B, Moy B, Isakoff SJ, Parmigiani G, Trippa L, Bardia A. Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: a comprehensive meta-analysis. Clin Cancer Res, 2020, 26(12): 2838-2848.
doi: 10.1158/1078-0432.CCR-19-3492 pmid: 32046998 |
| [11] | Rafique R, Islam SMR, Kazi JU. Machine learning in the prediction of cancer therapy. Comput Struct Biotechnol J, 2021, 19: 4003-4017. |
| [12] | Osama S, Shaban H, Ali AA. Gene reduction and machine learning algorithms for cancer classification based on microarray gene expression data: A comprehensive review. Expert Syst Appl, 2023, 213: 118946. |
| [13] | Yuan TW, Edelmann D, Fan ZW, Alwers E, Kather JN, Brenner H, Hoffmeister M. Machine learning in the identification of prognostic DNA methylation biomarkers among patients with cancer: A systematic review of epigenome-wide studies. Artif Intell Med, 2023, 143: 102589. |
| [14] | Lever J, Krzywinski M, Altman N. Logistic regression. Nat Methods, 2016, 13(7): 541-542. |
| [15] | Kourou K, Exarchos KP, Papaloukas C, Sakaloglou P, Exarchos T, Fotiadis DI. Applied machine learning in cancer research: A systematic review for patient diagnosis, classification and prognosis. Comput Struct Biotechnol J, 2021, 19: 5546-5555. |
| [16] | Audureau E, Soubeyran PL, Martinez-Tapia C, Bellera CA, Bastuji-Garin S, Boudou-Rouquette P, Rainfray M, Chahwakilian A, Grellety T, Hanon O, Mathoulin-Pélissier S, Paillaud E, Canoui-Poitrine F. Using machine learning to predict mortality in older patients with cancer: Decision tree and random forest analyses from the ELCAPA and ONCODAGE prospective cohorts. J Clin Oncol, 2019, 37(15_suppl): 11516. |
| [17] | Paul A, Mukherjee DP, Das P, Gangopadhyay A, Chintha AR, Kundu S. Improved random forest for classification. IEEE Trans Image Process, 2018, 27(8): 4012-4024. |
| [18] | Zolfaghari B, Mirsadeghi L, Bibak K, Kavousi K. Cancer prognosis and diagnosis methods based on ensemble learning. ACM Comput Surv, 2023, 55(12): 1-34. |
| [19] | Shahraki A, Abbasi M, Haugen Ø. Boosting algorithms for network intrusion detection: a comparative evaluation of real AdaBoost, gentle AdaBoost and modest AdaBoost. Eng Appl Artif Intell, 2020, 94: 103770. |
| [20] | Tseng CJ, Lu CJ, Chang CC, Chen GD, Cheewakriangkrai C. Integration of data mining classification techniques and ensemble learning to identify risk factors and diagnose ovarian cancer recurrence. Artif Intell Med, 2017, 78: 47-54. |
| [21] | Wickramasinghe I, Kalutarage H. Naive Bayes: applications, variations and vulnerabilities: a review of literature with code snippets for implementation. Soft Comput, 2021, 25(3): 2277-2293. |
| [22] | Höhn J, Krieghoff-Henning E, Jutzi TB, von Kalle C, Utikal JS, Meier F, Gellrich FF, Hobelsberger S, Hauschild A, Schlager JG, French L, Heinzerling L, Schlaak M, Ghoreschi K, Hilke FJ, Poch G, Kutzner H, Heppt MV, Haferkamp S, Sondermann W, Schadendorf D, Schilling B, Goebeler M, Hekler A, Fröhling S, Lipka DB, Kather JN, Krahl D, Ferrara G, Haggenmüller S, Brinker TJ. Combining CNN-based histologic whole slide image analysis and patient data to improve skin cancer classification. Eur J Cancer, 2021, 149: 94-101. |
| [23] | Zhang Y, Zhang C, Li KJ, Deng JL, Liu H, Lai GC, Xie B, Zhong XN. Identification of molecular subtypes and prognostic characteristics of adrenocortical carcinoma based on unsupervised clustering. Int J Mol Sci, 2023, 24(20): 15465. |
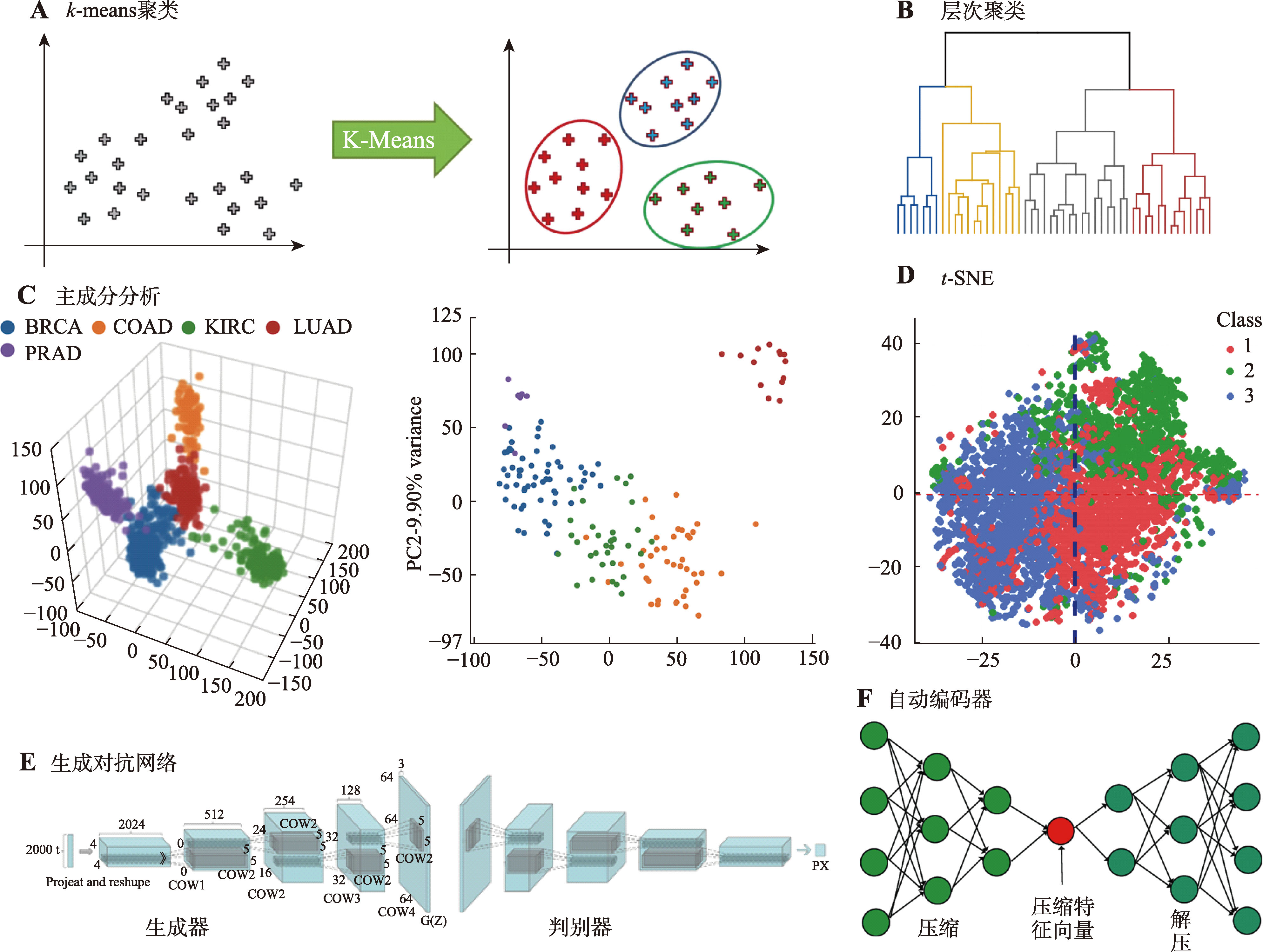
| [24] | Ayesha S, Hanif MK, Talib R. Overview and comparative study of dimensionality reduction techniques for high dimensional data. Inf Fusion, 2020, 59: 44-58. |
| [25] |
Nidheesh N, Abdul Nazeer KA, Ameer PM. An enhanced deterministic K-Means clustering algorithm for cancer subtype prediction from gene expression data. Comput Biol Med, 2017, 91: 213-221.
doi: S0010-4825(17)30340-2 pmid: 29100115 |
| [26] |
Wei W, Liang JY, Guo XY, Song P, Sun YJ. Hierarchical division clustering framework for categorical data. Neurocomputing, 2019, 341: 118-134.
doi: 10.1016/j.neucom.2019.02.043 |
| [27] | Levada ALM. Parametric PCA for unsupervised metric learning. Pattern Recognit Lett, 2020, 135: 425-430. |
| [28] | Haberl D, Spielvogel CP, Jiang ZW, Orlhac F, Iommi D, Carrió I, Buvat I, Haug AR, Papp L. Multicenter PET image harmonization using generative adversarial networks. Eur J Nucl Med Mol Imaging, 2024, 51(9): 2532-2546. |
| [29] | Saghand PG, Naqa IE, Tan AC, Xie MY, Dai DH, Chen JL, Ratan A, McCarter M, Carpten JD, Shah H, Ikeguchi A, Tripathi A, Puzanov I, Arnold SM, Churchman ML, Hwu P, Conejo-Garcia J, Dalton WS, Weiner GJ, Tarhini AA. A deep learning approach utilizing clinical and molecular data for identifying prognostic biomarkers in patients treated with immune checkpoint inhibitors: An ORIEN pan-cancer study. J Clin Oncol, 2022, 40(16_suppl): 2619. |
| [30] | Liao WJ, He JL, Luo XD, Wu MW, Shen YY, Li CR, Xiao JH, Wang GT, Chen NY. Automatic delineation of gross tumor volume based on magnetic resonance imaging by performing a novel semisupervised learning framework in nasopharyngeal carcinoma. Int J Radiat Oncol Biol Phys, 2022, 113(4): 893-902. |
| [31] | Guha S. LASSO based analysis for prediction of prognostic signature genes associated with breast cancer. bioRxiv, 2024: 587421. |
| [32] |
He JJ, Fu FM, Wang W, Xi GQ, Guo WH, Zheng LQ, Ren WJ, Qiu LD, Huang XX, Wang C, Li LH, Kang DY, Chen JX. Prognostic value of tumour-infiltrating lymphocytes based on the evaluation of frequency in patients with oestrogen receptor-positive breast cancer. Eur J Cancer, 2021, 154: 217-226.
doi: 10.1016/j.ejca.2021.06.011 pmid: 34293665 |
| [33] |
Joo S, Ko ES, Kwon S, Jeon E, Jung H, Kim JY, Chung MJ, Im YH. Multimodal deep learning models for the prediction of pathologic response to neoadjuvant chemotherapy in breast cancer. Sci Rep, 2021, 11(1): 18800.
doi: 10.1038/s41598-021-98408-8 pmid: 34552163 |
| [34] |
Tong L, Wu H, Wang MD. Integrating multi-omics data by learning modality invariant representations for improved prediction of overall survival of cancer. Methods, 2021, 189: 74-85.
doi: 10.1016/j.ymeth.2020.07.008 pmid: 32763377 |
| [35] | Palmal S, Arya N, Saha S, Tripathy S. Integrative prognostic modeling for breast cancer: Unveiling optimal multimodal combinations using graph convolutional networks and calibrated random forest. Appl Soft Comput, 2024, 154: 111379. |
| [36] |
Arya N, Saha S, Mathur A, Saha S. Improving the robustness and stability of a machine learning model for breast cancer prognosis through the use of multi-modal classifiers. Sci Rep, 2023, 13(1): 4079.
doi: 10.1038/s41598-023-30143-8 pmid: 36906618 |
| [37] | Jiang YZ, Ma D, Jin X, Xiao Y, Yu Y, Shi JX, Zhou YF, Fu T, Lin CJ, Dai LJ, Liu CL, Zhao S, Su GH, Hou WW, Liu YQ, Chen QW, Yang JC, Zhang NX, Zhang WJ, Liu W, Ge WG, Yang WT, You C, Gu YJ, Kaklamani V, Bertucci F, Verschraegen C, Daemen A, Shah NM, Wang T, Guo TN, Shi LM, Perou CM, Zheng YT, Huang W, Shao ZM. Integrated multiomic profiling of breast cancer in the Chinese population reveals patient stratification and therapeutic vulnerabilities. Nat Cancer, 2024, 5(4): 673-690. |
| [38] | Li X, Yang LF, Jiao X. Deep learning-based multiomics integration model for predicting axillary lymph node metastasis in breast cancer. Future Oncol, 2023, 19(20): 1429-1438. |
| [39] | Zhang LY, Pan J, Wang Z, Yang CH, Chen WZ, Jiang JX, Zheng ZY, Jia F, Zhang Y, Jiang JH, Su K, Ren GH, Huang J. Multi-omics profiling suggesting intratumoral mast cells as predictive index of breast cancer lung metastasis. Front Oncol, 2022, 11: 788778. |
| [40] | Masood S. Neoadjuvant chemotherapy in breast cancers. Womens Health (Lond), 2016, 12(5): 480-491. |
| [41] | Wheeler SB, Rotter J, Gogate A, Reeder-Hayes KE, Drier SW, Ekwueme DU, Fairley TL, Rocque GB, Trogdon JG. Cost-effectiveness of pharmacologic treatment options for women with endocrine-refractory or triple-negative metastatic breast cancer. J Clin Oncol, 2023, 41(1): 32-42. |
| [42] |
Hurvitz SA, Martin M, Symmans WF, Jung KH, Huang CS, Thompson AM, Harbeck N, Valero V, Stroyakovskiy D, Wildiers H, Campone M, Boileau JF, Beckmann MW, Afenjar K, Fresco R, Helms HJ, Xu J, Lin YG, Sparano J, Slamon D. Neoadjuvant trastuzumab, pertuzumab, and chemotherapy versus trastuzumab emtansine plus pertuzumab in patients with HER2-positive breast cancer (KRISTINE): a randomised, open-label, multicentre, phase 3 trial. Lancet Oncol, 2018, 19(1): 115-126.
doi: S1470-2045(17)30716-7 pmid: 29175149 |
| [43] | Thota R, Christensen B, Fulde G, Lewis MA, Haslem DS, Rhodes TD, Nadauld L, Barker T. Characterization of the tumor mutation burden in hepatobiliary tumors. J Clin Oncol, 2019, 37(4_suppl): 295. |
| [44] | Laas E, Labrosse J, Hamy AS, Benchimol G, de Croze D, Feron JG, Coussy F, Balezeau T, Guerin J, Lae M, Pierga JY, Reyal F. Determination of breast cancer prognosis after neoadjuvant chemotherapy: comparison of Residual Cancer Burden (RCB) and Neo-Bioscore. Br J Cancer, 2021, 124(8): 1421-1427. |
| [45] |
Walia A, Tuia J, Prasad V. Progression-free survival, disease-free survival and other composite end points in oncology: improved reporting is needed. Nat Rev Clin Oncol, 2023, 20(12): 885-895.
doi: 10.1038/s41571-023-00823-5 pmid: 37828154 |
| [46] | Wang H, Mao XY. Evaluation of the efficacy of neoadjuvant chemotherapy for breast cancer. Drug Des Devel Ther, 2020, 14: 2423-2433. |
| [47] |
Liu ZY, Li ZL, Qu JR, Zhang RZ, Zhou XZ, Li LF, Sun K, Tang ZC, Jiang H, Li HL, Xiong QQ, Ding YY, Zhao XM, Wang K, Liu ZY, Tian J. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: a multicenter study. Clin Cancer Res, 2019, 25(12): 3538-3547.
doi: 10.1158/1078-0432.CCR-18-3190 pmid: 30842125 |
| [48] | Park S, Yi G. Development of gene expression-based random forest model for predicting neoadjuvant chemotherapy response in triple-negative breast cancer. Cancers (Basel), 2022, 14(4): 881. |
| [49] | Sammut SJ, Crispin-Ortuzar M, Chin SF, Provenzano E, Bardwell HA, Ma WX, Cope W, Dariush A, Dawson SJ, Abraham JE, Dunn J, Hiller L, Thomas J, Cameron DA, Bartlett JMS, Hayward L, Pharoah PD, Markowetz F, Rueda OM, Earl HM, Caldas C. Multi-omic machine learning predictor of breast cancer therapy response. Nature, 2022, 601(7894): 623-629. |
| [50] |
Malik V, Kalakoti Y, Sundar D. Deep learning assisted multi-omics integration for survival and drug-response prediction in breast cancer. BMC Genomics, 2021, 22(1): 214.
doi: 10.1186/s12864-021-07524-2 pmid: 33761889 |
| [51] | Anderson NM, Simon MC. The tumor microenvironment. Curr Biol, 2020, 30(16): R921-R925. |
| [52] |
Mittal S, Brown NJ, Holen I. The breast tumor microenvironment: role in cancer development, progression and response to therapy. Expert Rev Mol Diagn, 2018, 18(3): 227-243.
doi: 10.1080/14737159.2018.1439382 pmid: 29424261 |
| [53] | Lucena-Sánchez E, Hicke FJ, Clara-Trujillo S, García- Fernández A, Martínez-Máñez R. Nanoparticle-mediated tumor microenvironment remodeling favors the communication with the immune cells for tumor killing. Adv Ther, 2024, 7(5): 2400004. |
| [54] | Keenan TE, Tolaney SM. Role of immunotherapy in triple-negative breast cancer. J Natl Compr Canc Netw, 2020, 18(4): 479-489. |
| [55] |
Shiravand Y, Khodadadi F, Kashani SMA, Hosseini-Fard SR, Hosseini S, Sadeghirad H, Ladwa R, O'Byrne K, Kulasinghe A. Immune checkpoint inhibitors in cancer therapy. Curr Oncol, 2022, 29(5): 3044-3060.
doi: 10.3390/curroncol29050247 pmid: 35621637 |
| [56] | Ambrosini M, Rousseau B, Manca P, Artz O, Marabelle A, André T, Maddalena G, Mazzoli G, Intini R, Cohen R, Cercek A, Segal NH, Saltz L, Varghese AM, Yaeger R, Nusrat M, Goldberg Z, Ku GY, El Dika I, Margalit O, Grinshpun A, Kasi PM, Schilsky R, Lutfi A, Shacham- Shmueli E, Khan Afghan M, Weiss L, Westphalen CB, Conca V, Decker B, Randon G, Elez E, Fakih M, Schrock AB, Cremolini C, Jayachandran P, Overman MJ, Lonardi S, Pietrantonio F. Immune checkpoint inhibitors for POLE or POLD1 proofreading-deficient metastatic colorectal cancer. Ann Oncol, 2024, 35(7): 643-655. |
| [57] |
Li YW, Wu X, Fang DY, Luo Y. Informing immunotherapy with multi-omics driven machine learning. NPJ Digit Med, 2024, 7(1): 67.
doi: 10.1038/s41746-024-01043-6 pmid: 38486092 |
| [58] |
Chen IX, Newcomer K, Pauken KE, Juneja VR, Naxerova K, Wu MW, Pinter M, Sen DR, Singer M, Sharpe AH, Jain RK. A bilateral tumor model identifies transcriptional programs associated with patient response to immune checkpoint blockade. Proc Natl Acad Sci USA, 2020, 117(38): 23684-23694.
doi: 10.1073/pnas.2002806117 pmid: 32907939 |
| [59] | Gou QH, Liu ZJ, Xie YX, Deng YL, Ma J, Li JP, Zheng H. Systematic evaluation of tumor microenvironment and construction of a machine learning model to predict prognosis and immunotherapy efficacy in triple-negative breast cancer based on data mining and sequencing validation. Front Pharmacol, 2022, 13: 995555. |
| [60] |
Xiao Y, Ma D, Zhao S, Suo C, Shi JX, Xue MZ, Ruan M, Wang H, Zhao JJ, Li Q, Wang P, Shi LM, Yang WT, Huang W, Hu X, Yu KD, Huang SL, Bertucci F, Jiang YZ, Shao ZM, AME Breast Cancer Collaborative Group. Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin Cancer Res, 2019, 25(16): 5002-5014.
doi: 10.1158/1078-0432.CCR-18-3524 pmid: 30837276 |
| [61] |
Hu QT, Hong Y, Qi P, Lu GQ, Mai XY, Xu S, He XY, Guo Y, Gao LL, Jing ZY, Wang JW, Cai T, Zhang Y. Atlas of breast cancer infiltrated B-lymphocytes revealed by paired single-cell RNA-sequencing and antigen receptor profiling. Nat Commun, 2021, 12(1): 2186.
doi: 10.1038/s41467-021-22300-2 pmid: 33846305 |
| [62] |
Wu SZ, Al-Eryani G, Roden DL, Junankar S, Harvey K, Andersson A, Thennavan A, Wang CF, Torpy JR, Bartonicek N, Wang TP, Larsson L, Kaczorowski D, Weisenfeld NI, Uytingco CR, Chew JG, Bent ZW, Chan CL, Gnanasambandapillai V, Dutertre CA, Gluch L, Hui MN, Beith J, Parker A, Robbins E, Segara D, Cooper C, Mak C, Chan B, Warrier S, Ginhoux F, Millar E, Powell JE, Williams SR, Liu XS, O’Toole S, Lim E, Lundeberg J, Perou CM, Swarbrick A. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet, 2021, 53(9): 1334-1347.
doi: 10.1038/s41588-021-00911-1 pmid: 34493872 |
| [63] | Jiménez-Santos MJ, García-Martín S, Rubio-Fernández M, Gómez-López G, Al-Shahrour F. Spatial transcriptomics in breast cancer reveals tumour microenvironment-driven drug responses and clonal therapeutic heterogeneity. bioRxiv, 2024, doi: 10.1101/2024.02.18.580660. |
| [64] | Hua Z, White J, Zhou JJ. Cancer stem cells in TNBC. Semin Cancer Biol, 2022, 82: 26-34. |
| [65] | Alaa AM, Gurdasani D, Harris AL, Rashbass J, van der Schaar M. Machine learning to guide the use of adjuvant therapies for breast cancer. Nat Mach Intell, 2021, 3(8): 716-726. |
| [1] | Hui Liang, Xue Wang, Jingfang Si, Yi Zhang. Classification accuracy of machine learning algorithms for Chinese local cattle breeds using genomic markers [J]. Hereditas(Beijing), 2024, 46(7): 530-539. |
| [2] | Huiyi Zheng, Huaxuan Wu, Zhiqiang Du. Gut metagenome-derived image augmentation and deep learning improve prediction accuracy of metabolic disease classification [J]. Hereditas(Beijing), 2024, 46(10): 886-896. |
| [3] | Yuxin Wan, Xinyu Zhu, Yu Zhao, Na Sun, Tiantongfei Jiang, Juan Xu. Computational dissection of the regulatory mechanisms of aberrant metabolism in remodeling the microenvironment of breast cancer [J]. Hereditas(Beijing), 2024, 46(10): 871-885. |
| [4] | Chunhui Ma, Haixu Hu, Lijuan Zhang, Yi Liu, Tianyi Liu. Establishment and verification of a digital PCR assay for the detection of CK19 expression in quantitative analysis of circulating tumor cell [J]. Hereditas(Beijing), 2023, 45(3): 250-260. |
| [5] | Dong Chang, Xiangxiang Liu, Rui Liu, Jianwei Sun. The role and regulatory mechanism of FSCN1 in breast tumorigenesis and progression [J]. Hereditas(Beijing), 2023, 45(2): 115-127. |
| [6] | Dong Chen, Shujie Wang, Zhenjian Zhao, Xiang Ji, Qi Shen, Yang Yu, Shengdi Cui, Junge Wang, Ziyang Chen, Jinyong Wang, Zongyi Guo, Pingxian Wu, Guoqing Tang. Genomic prediction of pig growth traits based on machine learning [J]. Hereditas(Beijing), 2023, 45(10): 922-932. |
| [7] | Yongqiang Kong, Jinkai Liu, Jiaqi Gu, Jingyi Xu, Yunuo Zheng, Yiliang Wei, Shaoyuan Wu. Optimization scheme of machine learning model for genetic division between northern Han, southern Han, Korean and Japanese [J]. Hereditas(Beijing), 2022, 44(11): 1028-1043. |
| [8] | Yali Hu, Rui Dai, Yongxin Liu, Jingying Zhang, Bin Hu, Chengcai Chu, Huaibo Yuan, Yang Bai. Analysis of rice root bacterial microbiota of Nipponbare and IR24 [J]. Hereditas(Beijing), 2020, 42(5): 506-518. |
| [9] | Qiang Zhang, Mingliang Gu. Single-cell sequencing and its application in breast cancer [J]. Hereditas(Beijing), 2020, 42(3): 250-268. |
| [10] | Xinyuan Wang, Yu Zhang, Nan Yang, He Cheng, Yujie Sun. DNMT3a mediates paclitaxel-induced abnormal expression of LINE-1 by increasing the intragenic methylation [J]. Hereditas(Beijing), 2020, 42(1): 100-111. |
| [11] | Qichao Yu,Bin Song,Xuanxuan Zou,Ling Wang,Dequan Liu,Bo Li,Kun Ma. Analysis of normal tissues adjacent to the tumour-specific expressed genes in breast cancer [J]. Hereditas(Beijing), 2019, 41(7): 625-633. |
| [12] | Tonglu YU,Dongliang Cai,Genfeng Zhu,Xiaojuan Ye,Taishan Min,Hongyan Chen,Daru Lu,Haoming Chen. Effects of CSN4 knockdown on proliferation and apoptosis of breast cancer MDA-MB-231 cells [J]. Hereditas(Beijing), 2019, 41(4): 318-326. |
| [13] | Zhao Xuetong, Yang Yadong, Qu Hongzhu, Fang Xiangdong. Applications of machine learning in clinical decision support in the omic era [J]. Hereditas(Beijing), 2018, 40(9): 693-703. |
| [14] | Zhang Guishan, Yang Yong, Zhang Lingmin, Dai Xianhua. Application of machine learning in the CRISPR/Cas9 system [J]. Hereditas(Beijing), 2018, 40(9): 704-723. |
| [15] | Zhe-ye Peng,Zi-jun Tang,Min-zhu Xie. Research progress in machine learning methods for gene-gene interaction detection [J]. Hereditas(Beijing), 2018, 40(3): 218-226. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||