遗传 ›› 2020, Vol. 42 ›› Issue (2): 172-182.doi: 10.16288/j.yczz.19-214
收稿日期:2019-10-02
修回日期:2019-12-13
出版日期:2020-02-20
发布日期:2019-12-23
基金资助:
Hong Xiang( ), Xiaohu Yang, Liangxia Ai, Yanping Pan, Yong Hu(
), Xiaohu Yang, Liangxia Ai, Yanping Pan, Yong Hu( )
)
Received:2019-10-02
Revised:2019-12-13
Published:2020-02-20
Online:2019-12-23
Supported by:摘要:
利用生物信息学方法分析脱发相关差异表达基因,有望帮助了解脱发发生发展的分子机制。本研究从NCBI的子数据库GEO中选择基因表达谱GSE45512和GSE45513数据集,利用R语言limma工具包,筛选出两个物种斑秃样本与正常样本的共同显著差异表达基因。对这部分基因进行功能注释和蛋白互作网络分析,同时对全部差异表达基因进行基因集富集分析。结果发现,人头皮斑秃样本共筛选出225个差异表达基因;C3H/HeJ小鼠自发斑秃皮肤样本共筛选出337个差异表达基因;两个物种的共同显著差异表达基因有23个。GO功能富集分析和蛋白互作网络分析显示,这部分差异基因显著富集于免疫相关功能,并且彼此间存在蛋白互作关系。基因集富集分析显示两个物种的差异基因都能显著富集到趋化因子信号通路、细胞因子受体相互作用、金葡菌感染及抗原加工与呈递通路;而且人的下调差异基因不仅映射到了人类表型数据库的脱发表型,也映射到皮肤附属物病理相关表型。综上所述,本研究通过生物信息方法分析脱发皮肤组织与正常皮肤组织的差异表达基因,最终筛选出23个在人和小鼠中共同存在的显著差异表达基因;此外,分析发现脱发与免疫过程及皮肤附属物病变密切相关,这些结果为脱发的诊断和治疗提供了新思路。
向虹, 阳小胡, 艾亮霞, 潘燕平, 胡勇. 脱发相关差异表达基因的生物信息学分析[J]. 遗传, 2020, 42(2): 172-182.
Hong Xiang, Xiaohu Yang, Liangxia Ai, Yanping Pan, Yong Hu. Bioinformatics analysis of differentially expressed genes on alopecia[J]. Hereditas(Beijing), 2020, 42(2): 172-182.
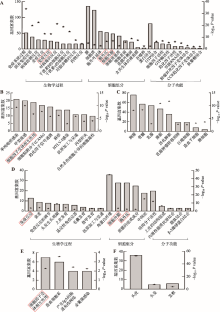
附图1
斑秃在人头皮样本和小鼠皮肤样本中的差异 利用DAVID功能注释工具分析人和小鼠中非共有差异表达基因。A:C3H/HeJ小鼠自发性AA皮肤非共有差异表达基因的GO注释分析,包括生物学过程(biological process),细胞组分(cellular component)和分子功能(molecular function)。B:C3H/HeJ小鼠自发性AA皮肤非共有差异表达基因所涉及的信号通路。C:C3H/HeJ小鼠自发性AA皮肤非共有差异表达基因的组织表达。D:人头皮非共有差异表达基因的GO注释分析。E:人头皮非共有差异表达基因所涉及的信号通路。F:人头皮非共有差异表达基因的组织表达。左侧Y轴代表所涉及的差异基因个数,用柱状图显示。右侧Y轴代表所涉及条目的显著性,用五角星显示,数据用-log10(P value)表示。A、B、D和E图的红色矩形框代表在人和鼠中共同出现的条目。"
| [1] |
Safavi KH, Muller SA, Suman VJ, Moshell AN, Melton LJ . Incidence of alopecia areata in olmsted county, minnesota, 1975 through 1989. Mayo Clin Proc, 1995,70(7):628-633.
doi: 10.4065/70.7.628 pmid: 7791384 |
| [2] |
Pratt CH, King LE Jr, Messenger AG, Christiano AM, Sundberg JP . Alopecia areata. Nat Rev Dis Primers, 2017,3:17011.
doi: 10.1038/nrdp.2017.11 pmid: 28300084 |
| [3] | Liu LY, King BA , Craiglow BG. Health-related quality of life(HRQoL)among patients with alopecia areata(AA): A systematic review. J Am Acad Dermatol, 2016, 75(4): 806-812.e3. |
| [4] |
Hunt N , McHale S. The psychological impact of alopecia. BMJ, 2005,331(7522):951-953.
doi: 10.1136/bmj.331.7522.951 pmid: 16239692 |
| [5] |
Singam V, Patel KR, Lee HH, Rastogi S, Silverberg JI . Association of alopecia areata with hospitalization for mental health disorders in US adults. J Am Acad Dermatol, 2019,80(3):792-794.
doi: 10.1016/j.jaad.2018.07.044 pmid: 30092332 |
| [6] |
Rencz F, Gulácsi L, Péntek M, Wikonkál N, Baji P, Brodszky V . Alopecia areata and health-related quality of life: a systematic review and meta-analysis. Br J Dermatol. 2016,175(3):561-571.
doi: 10.1111/bjd.14497 pmid: 26914830 |
| [7] |
Xing LZ, Dai ZP, Jabbari A, Cerise JE, Higgins CA , Gong WJ, de Jong A, Harel S, DeStefano GM, Rothman L, Singh P, Petukhova L, Mackay-Wiggan J, Christiano AM, Clynes R. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat Med, 2014,20(9):1043-1049.
doi: 10.1038/nm.3645 pmid: 25129481 |
| [8] |
The Gene Ontology Consortium . The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res, 2019,47(D1):D330-d338.
doi: 10.1093/nar/gky1055 pmid: 30395331 |
| [9] |
Huang da W, Sherman BT, Lempicki RA . Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res, 2009,37(1):1-13.
doi: 10.1093/nar/gkn923 pmid: 19033363 |
| [10] |
Chen HA, Pan JY, Chiang CH, Jhang AH, Ho WT . New idea for hair transplantation to preserve more donor hair follicles. Med Hypotheses, 2019,128:83-85.
doi: 10.1016/j.mehy.2019.05.018 pmid: 31203916 |
| [11] |
Afifi L, Maranda EL, Zarei M, Delcanto GM, Falto- Aizpurua L, Kluijfhout WP, Jimenez JJ . Low-level laser therapy as a treatment for androgenetic alopecia. Lasers Surg Med, 2017,49(1):27-39.
doi: 10.1002/lsm.22512 pmid: 27114071 |
| [12] |
Tekin NS, Tekin IO, Cinar S, Altinyazar HC, Koca R, Esturk E . The PUVA-turban as an alternative treatment of alopecia associated with chronic graft versus host disease. J Am Acad Dermatol, 2005,53(5):902-903.
doi: 10.1016/j.jaad.2005.04.078 pmid: 16243155 |
| [13] |
Lee HW, Jun JH, Lee JA, Lim HJ, Lim HS, Lee MS . Acupuncture for treating alopecia areata: a protocol of systematic review of randomised clinical trials. BMJ Open, 2015,5(10):e008841.
doi: 10.1136/bmjopen-2015-008841 pmid: 26503391 |
| [14] |
Suchonwanit P, Srisuwanwattana P, Chalermroj N, Khunkhet S . A randomized, double-blind controlled study of the efficacy and safety of topical solution of 0.25% finasteride admixed with 3% minoxidil vs. 3% minoxidil solution in the treatment of male androgenetic alopecia. J Eur Acad Dermatol Venereol, 2018,32(12):2257-2263.
doi: 10.1111/jdv.15171 pmid: 29972712 |
| [15] |
Durdu M, Özcan D, Baba M, Seçkin D . Efficacy and safety of diphenylcyclopropenone alone or in combination with anthralin in the treatment of chronic extensive alopecia areata: a retrospective case series. J Am Acad Dermatol, 2015,72(4):640-650.
doi: 10.1016/j.jaad.2015.01.008 pmid: 25653027 |
| [16] |
Rodrigues BL, Montalvão SAL, Cancela RBB, Silva FAR, Urban A, Huber SC, Júnior JLRC, Lana JFSD, Annichinno- Bizzacchi JM . Treatment of male pattern alopecia with platelet-rich plasma: A double-blind controlled study with analysis of platelet number and growth factor levels. J Am Acad Dermatol, 2019,80(3):694-700.
doi: 10.1016/j.jaad.2018.09.033 pmid: 30287324 |
| [17] |
Sundberg JP, Boggess D, Montagutelli X, Hogan ME, King LE Jr . C3H/HeJ mouse model for alopecia areata. J Invest Dermatol, 1995,104(5 Suppl):16s-17s.
doi: 10.1038/jid.1995.38 pmid: 7738375 |
| [18] |
Subramanya RD, Coda AB, Sinha AA . Transcriptional profiling in alopecia areata defines immune and cell cycle control related genes within disease-specific signatures. Genomics, 2010,96(3):146-153.
doi: 10.1016/j.ygeno.2010.05.002 pmid: 20546884 |
| [19] |
Kawano M, Umeda S, Yasuda T, Fujita M, Ishikawa A, Imamura T, Imai T, Nakayama F . FGF18 signaling in the hair cycle resting phase determines radioresistance of hair follicles by arresting hair cycling. Adv Radiat Oncol, 2016,1(3):170-181.
doi: 10.1016/j.adro.2016.05.004 pmid: 28740887 |
| [20] |
Midorikawa T, Chikazawa T, Yoshino T, Takada K, Arase S . Different gene expression profile observed in dermal papilla cells related to androgenic alopecia by DNA macroarray analysis. J Dermatol Sci, 2004,36(1):25-32.
doi: 10.1016/j.jdermsci.2004.05.001 pmid: 15488702 |
| [21] |
Plikus MV, Widelitz RB, Maxson R, Chuong CM . Analyses of regenerative wave patterns in adult hair follicle populations reveal macro-environmental regulation of stem cell activity. Int J Dev Biol, 2009,53(5-6):857-868.
doi: 10.1387/ijdb.072564mp pmid: 19378257 |
| [22] |
Renninger ML, Seymour RE, Whiteley LO, Sundberg JP, Hogenesch H . Anti-IL5 decreases the number of eosinophils but not the severity of dermatitis in Sharpin-deficient mice. Exp Dermatol, 2010,19(3):252-258.
doi: 10.1111/j.1600-0625.2009.00944.x pmid: 19650867 |
| [23] |
Duncan FJ, Silva KA, Johnson CJ, King BL, Szatkiewicz JP, Kamdar SP, Ong DE, Napoli JL, Wang J, King LE Jr, Whiting DA , McElwee KJ, Sundberg JP, Everts HB. Endogenous retinoids in the pathogenesis of alopecia areata. J Invest Dermatol, 2013,133(2):334-343.
doi: 10.1038/jid.2012.344 pmid: 23014334 |
| [24] | Zainodini N, Hassanshahi G, Arababadi MK, Khorramdelazad H, Mirzaei A . Differential expression of CXCL1, CXCL9, CXCL10 and CXCL12 chemokines in alopecia areata. Iran J Immunol, 2013,10(1):40-46. |
| [25] |
Maouia A, Sormani L, Youssef M, Helal AN, Kassab A, Passeron T . Differential expression of CXCL9, CXCL10, and IFN-γ in vitiligo and alopecia areata patients. Pigment Cell Melanoma Res, 2017,30(2):259-261.
doi: 10.1111/pcmr.12559 pmid: 27863059 |
| [26] |
Bilgic O, Sivrikaya A, Unlu A, Altinyazar HC . Serum cytokine and chemokine profiles in patients with alopecia areata. J Dermatolog Treat, 2016,27(3):260-263.
doi: 10.3109/09546634.2015.1093591 pmid: 26367497 |
| [27] |
McPhee CG, Duncan FJ, Silva KA, King LE Jr, Hogenesch H, Roopenian DC, Everts HB, Sundberg JP . Increased expression of Cxcr3 and its ligands, Cxcl9 and Cxcl10, during the development of alopecia areata in the mouse. J Invest Dermatol, 2012,132(6):1736-1738.
doi: 10.1038/jid.2012.17 pmid: 22358057 |
| [28] |
Suárez-Fariñas M, Ungar B, Noda S, Shroff A, Mansouri Y, Fuentes-Duculan J, Czernik A, Zheng X, Estrada YD, Xu H, Peng X, Shemer A, Krueger JG, Lebwohl MG, Guttman-Yassky E . Alopecia areata profiling shows TH1, TH2, and IL-23 cytokine activation without parallel TH17/ TH22 skewing. J Allergy Clin Immunol, 2015,136(5):1277-1287.
doi: 10.1016/j.jaci.2015.06.032 pmid: 26316095 |
| [29] |
Dai Z, Xing L, Cerise J, Wang EH , Jabbari A, de Jong A, Petukhova L, Christiano AM, Clynes R. CXCR3 blockade inhibits T Cell migration into the skin and prevents development of alopecia areata. J Immunol, 2016,197(4):1089-1099.
doi: 10.4049/jimmunol.1501798 pmid: 27412416 |
| [30] |
Ito T, Hashizume H, Shimauchi T, Funakoshi A, Ito N, Fukamizu H, Takigawa M, Tokura Y . CXCL10 produced from hair follicles induces Th1 and Tc1 cell infiltration in the acute phase of alopecia areata followed by sustained Tc1 accumulation in the chronic phase. J Dermatol Sci, 2013,69(2):140-147.
doi: 10.1016/j.jdermsci.2012.12.003 pmid: 23312578 |
| [31] |
Gilhar A, Schrum AG, Etzioni A, Waldmann H, Paus R . Alopecia areata: Animal models illuminate autoimmune pathogenesis and novel immunotherapeutic strategies. Autoimmun Rev, 2016,15(7):726-735.
doi: 10.1016/j.autrev.2016.03.008 pmid: 26971464 |
| [32] |
Strazzulla LC, Wang EHC, Avila L, Lo Sicco K, Brinster N, Christiano AM, Shapiro J . Alopecia areata: Disease characteristics, clinical evaluation, and new perspectives on pathogenesis. J Am Acad Dermatol, 2018,78(1):1-12.
doi: 10.1016/j.jaad.2017.04.1141 pmid: 29241771 |
| [33] |
Liu LY, Craiglow BG, Dai F, King BA . Tofacitinib for the treatment of severe alopecia areata and variants: A study of 90 patients. J Am Acad Dermatol, 2017,76(1):22-28.
doi: 10.1016/j.jaad.2016.09.007 pmid: 27816293 |
| [34] |
Mackay-Wiggan J, Jabbari A, Nguyen N, Cerise JE, Clark C, Ulerio G, Furniss M, Vaughan R, Christiano AM, Clynes R . Oral ruxolitinib induces hair regrowth in patients with moderate-to-severe alopecia areata. JCI Insight, 2016,1(15):e89790.
doi: 10.1172/jci.insight.89790 pmid: 27699253 |
| [35] |
Jabbari A, Dai Z, Xing L, Cerise JE, Ramot Y, Berkun Y, Sanchez GA, Goldbach-Mansky R, Christiano AM, Clynes R, Zlotogorski A . Reversal of alopecia areata following treatment with the JAK1/2 inhibitor baricitinib. EBioMedicine, 2015,2(4):351-355.
doi: 10.1016/j.ebiom.2015.02.015 pmid: 26137574 |
| [36] |
Chelidze K, Lipner SR . Nail changes in alopecia areata: an update and review. Int J Dermatol, 2018,57(7):776-783.
doi: 10.1111/ijd.13866 pmid: 29318582 |
| [37] |
Kasumagic-Halilovic E, Prohic A . Nail changes in alopecia areata: frequency and clinical presentation. J Eur Acad Dermatol Venereol, 2009,23(2):240-241.
doi: 10.1111/j.1468-3083.2008.02830.x pmid: 18540984 |
| [38] |
Hamed FN , McDonagh AJG, Almaghrabi S, Bakri Y, Messenger AG, Tazi-Ahnini R. Epigallocatechin-3 gallate inhibits STAT-1/JAK2/IRF-1/HLA-DR/HLA-B and reduces CD8 MKG2D lymphocytes of alopecia areata patients. Int J Environ Res Public Health, 2018,15(12).
doi: 10.3390/ijerph15122882 pmid: 30558329 |
| [39] |
Shi JX, Peng P, Liu WX, Mi P, Xing C, Ning GZ, Feng SQ . Bioinformatics analysis of genes associated with the patchy-type alopecia areata: CD2 may be a new therapeutic target. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub, 2019, 1-7.
doi: 10.5507/bp.2019.049 pmid: 31558844 |
| [40] |
Nagao K, Kobayashi T, Moro K, Ohyama M, Adachi T, Kitashima DY, Ueha S, Horiuchi K, Tanizaki H, Kabashima K, Kubo A, Cho YH, Clausen BE, Matsushima K, Suematsu M, Furtado GC, Lira SA, Farber JM, Udey MC, Amagai M . Stress-induced production of chemokines by hair follicles regulates the trafficking of dendritic cells in skin. Nat Immunol, 2012,13(8):744-752.
doi: 10.1038/ni.2353 pmid: 22729248 |
| [41] |
Ibusuki A, Nishikawa T, Hiraki T, Okano T, Imai K, Kanegane H, Ohnishi H, Kato Z, Fujii K, Tanimoto A, Kawano Y, Kanekura T . Prominent dermal Langerhans cells in an Omenn syndrome patient with a novel mutation in the IL2RG gene. J Dermatol, 2019,46(11):1019-1023.
doi: 10.1111/1346-8138.15054 pmid: 31456262 |
| [42] |
Jiang W, Fan YX, Qiao X, Zhang YJ, Liu ZH, Zhao YH, Wang RJ, Wang ZX, Zhang WG, Su R, Li JQ . The transcriptome research progresses of skin hair follicle development. Hereditas(Beijing), 2015,37(6):528-534.
doi: 10.16288/j.yczz.14-440 pmid: 26351048 |
|
江玮, 范一星, 乔贤, 张燕军, 刘志红, 赵艳红, 王瑞军, 王志新, 张文广, 苏蕊, 李金泉 . 皮肤毛囊发育的转录组研究进展. 遗传, 2015,37(6):528-534.
doi: 10.16288/j.yczz.14-440 pmid: 26351048 |
| [1] | 陈瑶, 温馨, 袁芳媛, 彭钞灵, 王翠喆, 张君, 孟平平. 基于生物信息学对SLC25A21下游靶基因的筛选及验证[J]. 遗传, 2024, 46(12): 1055-1065. |
| [2] | 温馨, 梅锦, 钱美玉, 蒋一丹, 王娟, 许士博, 王翠喆, 张君. 基于转录组测序对GULP1下游靶基因筛选及分析[J]. 遗传, 2024, 46(10): 860-870. |
| [3] | 王姗姗, 赵琬怡, 吴慧潇, 舒梦, 袁嘉欣, 方丽, 徐潮. 特发性低促性腺激素性性腺功能减退症FGFR1与CEP290基因变异研究[J]. 遗传, 2022, 44(10): 937-949. |
| [4] | 吕红强, 郝乐乐, 刘二虎, 吴志芳, 韩九强, 刘源. 基于生物信息学的Hi-C研究现状与发展趋势[J]. 遗传, 2020, 42(1): 87-99. |
| [5] | 卢涣滋,王迪侃,王智. HPV阳性口咽癌患者预后与T细胞浸润和新抗原负荷相关性分析[J]. 遗传, 2019, 41(8): 725-735. |
| [6] | 张源笙,夏琳,桑健,李漫,刘琳,李萌伟,牛广艺,曹佳宝,滕徐菲,周晴,章张. 生命与健康大数据中心资源[J]. 遗传, 2018, 40(11): 1039-1043. |
| [7] | 向小华, 吴新儒, 晁江涛, 杨明磊, 杨帆, 陈果, 刘贯山, 王元英. 普通烟草WRKY基因家族的鉴定及表达分析[J]. 遗传, 2016, 38(9): 840-856. |
| [8] | 李晓旭, 刘成, 李伟, 张增林, 高晓明, 周慧, 郭永峰. 番茄WOX转录因子家族的鉴定及其进化、表达分析[J]. 遗传, 2016, 38(5): 444-460. |
| [9] | 周学, 杜宜兰, 金萍, 马飞. 癌症相关microRNA与靶基因的生物信息学分析[J]. 遗传, 2015, 37(9): 855-864. |
| [10] | 方翔, 李宁求, 付小哲, 李凯彬, 林强, 刘礼辉, 石存斌, 吴淑勤. 基于“天河二号”的水产病原生物信息分析平台构建及其在水产病原分析中的应用[J]. 遗传, 2015, 37(7): 702-710. |
| [11] | 邱红梅,郝文媛,高淑芹,马晓萍,郑宇宏,孟凡凡,范旭红,王洋,王跃强,王曙明. 大豆含硫氨基酸相关酶基因发掘[J]. 遗传, 2014, 36(9): 934-942. |
| [12] | 施杨, 徐筱, 李昊阳, 徐倩, 徐吉臣. 水稻扩展蛋白家族的生物信息学分析[J]. 遗传, 2014, 36(8): 809-820. |
| [13] | 齐鲁, 丁彦青. CREB5在大肠癌转移中的调控机制[J]. 遗传, 2014, 36(7): 679-684. |
| [14] | 陈先知, 王燕, 史建磊, 朱隆静, 王克磊, 徐坚. 黄瓜全基因组热激转录因子(HSFs)的鉴定与表达分析[J]. 遗传, 2014, 36(4): 376-386. |
| [15] | 程燕, 陈琳, 曹忻, 哈斯其美格, 谢小冬. Hsa-miR-125b在人胃癌耐药细胞株中的表达及其靶基因的功能分析[J]. 遗传, 2014, 36(2): 119-126. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
