遗传 ›› 2020, Vol. 42 ›› Issue (5): 423-434.doi: 10.16288/j.yczz.19-354
• 综述 • 下一篇
收稿日期:2020-01-07
修回日期:2020-03-24
出版日期:2020-05-20
发布日期:2020-04-01
通讯作者:
李利,张红平
E-mail:lily@sicau.edu.cn;zhp@sicau.edu.cn
基金资助:
Shuailong Zheng, Li Li( ), Hongping Zhang(
), Hongping Zhang( )
)
Received:2020-01-07
Revised:2020-03-24
Online:2020-05-20
Published:2020-04-01
Contact:
Li Li,Zhang Hongping
E-mail:lily@sicau.edu.cn;zhp@sicau.edu.cn
Supported by:摘要:
随着高通量测序技术和翻译组学研究的快速发展,对环状RNA (circular RNA, circRNA)翻译能力的研究日益成为热点。已有研究表明,circRNA自身可以翻译为蛋白,其蛋白功能与人类疾病发生发展有着密切联系,而且其有望成为mRNA的理想替代品,未来可被广泛地应用在蛋白质工程。本文系统综述了circRNA来源、形成方式和主要特征、翻译蛋白的方式、翻译能力的鉴定和功能验证,归纳了近年来circRNA翻译在人类疾病中的研究进展及其在蛋白质工程的应用,并对后续研究关注的问题进行了展望,以期为相关领域的研究提供参考。
郑帅龙, 李利, 张红平. 环状RNA翻译能力研究进展[J]. 遗传, 2020, 42(5): 423-434.
Shuailong Zheng, Li Li, Hongping Zhang. Progress on translation ability of circular RNA[J]. Hereditas(Beijing), 2020, 42(5): 423-434.
图1
circRNA的环化方式 A:套索驱动环化。当mRNA前体(pre-mRNA)进行GU/AG剪切时,可以跨外显子进行剪切,形成含有外显子和内含子的套索中间体,然后加工形成circRNA。B:内含子套索驱动环化。一些内含子的5?端附近含有7 nt的GU富集序列,3?端附近含有11 nt的C富集序列,它们可以在RNA polymerase II作用下形成一个套索内含子,进一步剪切形成ciRNA。C:内含子配对驱动环化。某些外显子两侧的内含子序列中含有反向互补序列,如RCMs (reverse complementary matches)、ICSs (intronic complementary sequences)及Alu元件,它们之间互补配对形成RNA双链,促进circRNA的形成。D:RNA结合蛋白驱动环化。一些RNA结合蛋白可以结合到外显子侧翼的内含子序列上,促进circRNA的形成。"
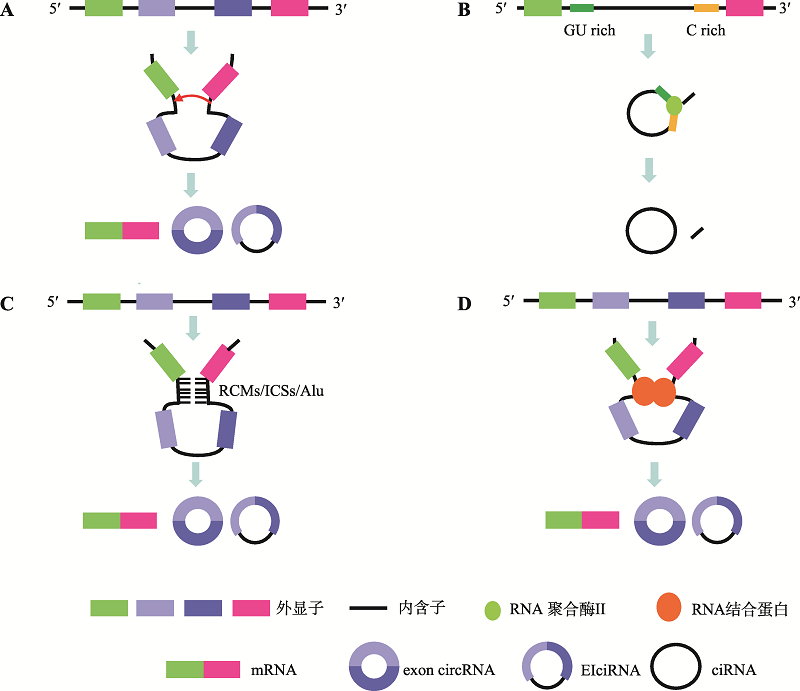
表1
研究circRNA翻译能力常用的预测工具"
| 名称 | 工具介绍 | 参考文献 |
|---|---|---|
| ORFfinder | 一个图形序列分析工具,可以分析并检索出circRNA序列中所有潜在的ORF | [39] |
| CPC2 | 通过输入circRNA序列,可预测其编码能力 | [40] |
| CPAT | 通过对circRNA序列信息进行分析,评估其编码潜力 | [41] |
| IRESfinder | 基于k-mer特征识别真核细胞中RNA序列上的IRES | [42] |
| IRESite | 基于已有68个病毒和115个真核细胞的实验数据,收录了大量的具有IRES位点的基因, 可对circRNA序列进行比对,预测circRNA上潜在的IRES位点 | [43] |
| Sramp | 对哺乳动物的circRNA序列进行检索,寻找m6A 基序 | [44] |
| m6Apred | 对酿酒酵母中m6A基序进行预测 | [45] |
| Pfam 32.0 | 通过预测circRNA翻译产物的潜在结构域,进而预测其功能 | [46] |
| CircCode | 基于RNA-seq或者Ribo-seq (核糖体印记与深度测序)数据,筛选结合有核糖体的circRNA | [47] |
表2
研究circRNA翻译能力常用的数据库"
| 名称 | 数据库介绍 | 参考文献 |
|---|---|---|
| circRNAdb | 收集了32,914条人类外显子circRNA记录,是首个汇总可编码蛋白的circRNA的数据库 | [48] |
| circBank | 收录了140,790条人类circRNA的记录,是目前关于人类已知circRNA最全的数据库 | [49] |
| circBase | 收集了包括人、小鼠、秀丽线虫(Caenorhabditis elegans)等共6个物种的circRNA信息,是目前涵盖物种数目最多的circRNA数据库 | [50] |
| CSCD | 肿瘤特异性circRNA数据库,可以对circRNA的RBP、ORF、亚细胞定位和可变剪切进行预测 | [51] |
| TSCD | circRNA组织特异性表达分析数据库,收录了人和小鼠的circRNA信息 | [52] |
| CIRCpedia V2 | 收录6个不同物种的180多个RNA-seq数据和26万个circRNA注释,可对人类和小鼠间circRNA进行保守分析 | [53] |
| CircInteractome | 通过预测circRNA是否含有已被验证过的IRES,判断其是否具有潜在的翻译能力 | [54] |
| CircNet | 对人类464个样本的RNA-seq数据系统地整理挖掘后建立的数据库,它提供了新circRNA的鉴定、circRNA-miRNA-mRNA的互作网络、circRNA亚型的表达水平、circRNA亚型的基因组注释和circRNA亚型的序列等信息 | [55] |
| plantcircbase | 植物circRNA的综合数据库,包含了拟南芥(Arabidopsis thaliana)、水稻(Oryza sativa)、玉米(Zea mays)、番茄(Solanum lycopersicum)和大麦(Hordeum vulgare)等物种的circRNA信息,可以使circRNA结构可视化 | [56] |
| Circ2Traits | 收录了与疾病和性状潜在相关的circRNA信息 | [57] |
| circAtlas | 收录了包括人、猕猴(Macaca mulatta)、小鼠、褐家鼠(Rattus norvegicus)、野猪(Sus scrofa)和鸡(Gallus domesticus)等物种的circRNA序列信息,可对不同物种间circRNA保守性进行分析 | [58] |
表3
研究circRNA翻译常用的实验方法"
| 方法 | 原理 | 用途 | 参考文献 |
|---|---|---|---|
| 核糖体印记与 深度测序技术 | 被核糖体包裹的RNA片段不易被RNase降解,回收片段测序得到与核糖体结合的序列信息 | 筛选出结合有核糖体的circRNA | [59,60] |
| 免疫组化反应 | 利用抗原与相应的抗体进行免疫反应后,通过相应的显色反应,以检测相应的目的蛋白的表达 | 检测circRNA生成的蛋白, 进行定性、定量和定位 | [61] |
| Western Blot | 设计特异性抗体与抗原进行免疫反应后,通过相应的显色反应,以检测相应的目的蛋白的表达 | 检测circRNA生成的蛋白,进行定性和定量 | [12] |
| 免疫沉淀 | 利用抗体特异性反应纯化富集目的蛋白 | 纯化富集目的蛋白 | [61] |
| 质谱检测 | 质谱分析是先将物质离子化,按离子的质荷比分离,然后测量各种离子谱峰的强度而实现分析目的的一种分析方法 | 检测circRNA新生成的蛋白,获得蛋白序列信息 | [61] |
| RNA结合蛋 白免疫沉淀 | 运用针对目标蛋白的抗体把相应的RNA-蛋白复合物沉淀下来,经过分离纯化就可以对结合在复合物上的RNA进行q-PCR验证或者测序分析 | 验证circRNA与翻译起始相关蛋白或因子的相互作用 | [62] |
| 蛋白质体外 结合实验 | 将RNA进行标记(如生物素探针标记),再与细胞裂解液共同孵育,从而形成RNA-蛋白质复合物,之后再分离复合物得到蛋白质,通过western blot或质谱检测检测蛋白质 | 验证circRNA与翻译起始相关蛋白或因子的相互作用 | [63] |
表4
circRNA翻译在人类疾病中的研究进展"
| circRNA | 蛋白 | 翻译蛋白方式 | 人类疾病 | 蛋白功能 | 参考文献 |
|---|---|---|---|---|---|
| circZNF609 | 未知 | IRES介导 | 杜氏型肌营养不良症 | 未知 | [12] |
| circFBXW7 | FBXW7-185aa | IRES介导 | 恶性胶质瘤 | 抑制 | [14] |
| circE7 | E7蛋白 | m6A介导 | 宫颈癌 | 促进 | [36] |
| circAKT3 | AKT3-174aa | IRES介导 | 恶性胶质瘤 | 抑制 | [61] |
| circPINT | PINT-87aa | IRES介导 | 恶性胶质瘤 | 抑制 | [64] |
| circβ-catenin | β-catenin-370aa | IRES介导 | 肝癌 | 促进 | [65] |
| circGprc5a | circGprc5a-peptide | 未知 | 膀胱癌 | 促进 | [66] |
| CDR1as、circANKH、 circASXL1、circCSNK1G3、 circKIAA1429和circSIPA1L1 | 未知 | 未知 | 扩张型心肌病 | 未知 | [67] |
| circSHPRH | SHPRH-146aa | IRES介导 | 恶性胶质瘤 | 抑制 | [68] |
| [1] | Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK . Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA, 1976,73(11):3852-3856. |
| [2] |
Grabowski PJ, Zaug AJ, Cech TR . The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of tetrahymena. Cell, 1981,23(2):467-476.
doi: 10.1016/0092-8674(81)90142-2 |
| [3] | Kjems J, Garrett RA . Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium desulfurococcus mobilis. Cell, 1988,54(5):693-703. |
| [4] | Zaphiropoulos PG . Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol, 1997,17(6):2985-2993. |
| [5] | Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R . Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell, 1993,73(5):1019-1030. |
| [6] | Zaphiropoulos PG . Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci USA, 1996,93(13):6536-6541. |
| [7] | Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (δ) virus possesses a circular RNA. Nature, 1986,323(6088):558-560. |
| [8] | Chen CY, Sarnow P . Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science, 1995,268(5209):415-417. |
| [9] | Perriman R, Ares M . Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA, 1998,4(9):1047. |
| [10] | Wang Y, Wang Z . Efficient backsplicing produces translatable circular mRNAs. RNA, 2015,21(2):172-179. |
| [11] | Pamudurti NR, Bartok O, Jens M, Ashwalfluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perezhernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S. Translation of circRNAs. Mol Cell, 2017, 66(1): 9-21.e7. |
| [12] | Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I . Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell, 2017,66(1):22-37. |
| [13] | Yang Y, Fan XJ, Mao MW, Song XW, Wu P, Zhang Y, Jin YF, Yang Y, Chen LL, Wang Y, Wong CC, Xiao XS, Wang ZF . Extensive translation of circular RNAs driven by N 6-methyladenosine . Cell Res, 2017,27(5):626-641. |
| [14] | Yang YB, Gao XY, Zhang ML, Yan S, Sun CJ, Xiao FZ, Huang NN, Yang XS, Zhao K, Zhou HK . Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer I, 2018,110(3):304-315. |
| [15] | Gao Y, Wang JF, Zheng Y, Zhang JY, Chen S, Zhao FQ . Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun, 2016,7:12060. |
| [16] | Chen LL, Yang L . Regulation of circRNA biogenesis. RNA Biol, 2015,12(4):381-388. |
| [17] | Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL . Circular intronic long noncoding RNAs. Mol Cell, 2013,51(6):792-806. |
| [18] | Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ . The RNA binding protein quaking regulates formation of circRNAs. Cell, 2015,160(6):1125-1134. |
| [19] | Suzuki H, Tsukahara T . A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci, 2014,15(6):9331-9342. |
| [20] | Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu JZ, Marzluff WF, Sharpless NE . Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA, 2013,19(2):141-157. |
| [21] | Venø MT, Hansen TB, Venø ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J . Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol, 2015,16(1):245. |
| [22] | Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N . Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell, 2015,58(5):870-885. |
| [23] | Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J . Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol, 2015,16(1):126. |
| [24] | You XT, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang MT, Glock C, Quedenau C, Wang X, Hou JY, Liu HY, Sun W, Sambandan S, Chen T, Schuman EM, Chen W . Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci, 2015,18(4):603-610. |
| [25] | Li ZY, Huang C, Bao C, Chen L, Lin M, Wang XL, Zhong GL, Yu B, Hu WC, Dai LM, Zhu PF, Chang ZX, Wu QF, Zhao Y, Jia Y, Xu P, Liu HJ, Shan G . Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol, 2015,22(3):256-264. |
| [26] | Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta C, Conn SJ . A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants, 2017,3(5):17053. |
| [27] | Xu HY, Guo S, Li W, Yu P . The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep, 2015,5(1):12453. |
| [28] |
Li H, Wei XF, Yang JM, Dong D, Hao D, Huang YZ, Lan XY, Plath M, Lei CZ, Ma Y, Lin FP, Bai YY, Chen H . CircFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Mol Ther Nucleic Acids, 2018,11:272-283.
doi: 10.1016/j.omtn.2018.02.012 |
| [29] | Feng Y, Hu AP, Li D, Wang JQ, Guo YH, Liu Y, Li HJ, Chen YJ, Wang XJ, Huang K, Zheng LD, Tong QS . Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer, 2019,18(1):158. |
| [30] | Dong R, Zhang XO, Zhang Y, Ma XK, Chen LL, Yang L . CircRNA-derived pseudogenes. Cell Res, 2016,26(6):747-750. |
| [31] | Chen GW, Shi YT, Liu MM, Sun JY. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis, 2018,9(2):175. |
| [32] | Haimov O, Sinvani H, Dikstein R . Cap-dependent, scanning-free translation initiation mechanisms. Biochim Biophys Acta, 2015,1849(11):1313-1318. |
| [33] | Liu XQ, Gao YB, Zhao LZ, Cai YC, Wang HY, Miao M, Gu LF, Zhang HX . Biogenesis, research methods, and functions of circular RNAs. Hereditas (Beijing), 2019,41(6):469-485. |
| 刘旭庆, 高宇帮, 赵良真, 蔡宇晨, 王汇源, 苗苗, 顾连峰, 张航晓 . 环状RNA的产生、研究方法及功能. 遗传, 2019,41(6):469-485. | |
| [34] | Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, Abe H . Rolling circle translation of circular RNA in living human cells. Sci Rep, 2015,5:16435. |
| [35] |
Ruggero D, Sonenberg N . The Akt of translational control. Oncogene, 2005,24(50):7426-7434.
doi: 10.1038/sj.onc.1209098 |
| [36] | Zhao JW, Lee EE, Kim J, Yang R, Chamseddin B, Ni CY, Gusho E, Xie Y, Chiang CM, Buszczak M, Zhan XW, Laimins L, Wang RC . Transforming activity of an oncoprotein-encoding circular RNA from human papillomavirus. Nat Commun, 2019,10(1):2300. |
| [37] | Zheng Y, Ji PF, Chen S, Hou LL, Zhao FQ. Reconstruction of full-length circular RNAs enables isoform-level quantification. Genome Med, 2019(1), 11:2. |
| [38] | Zhang JY, Chen S, Yang JW, Zhao FQ . Accurate quantification of circular RNAs identififies extensive circular isoform switching events. Nat Commun, 2020,11:90. |
| [39] |
Stothard P . The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques, 2000,28(6):1102-1104.
doi: 10.2144/00286ir01 |
| [40] | Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei LP, Gao G . CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res, 2007,35:345-349. |
| [41] |
Wang LG, Park HJ, Dasari S, Wang SQ, Kocher JP, Li W . CPAT: coding-potential assessment tool using an alignment- free logistic regression model. Nucleic Acids Res, 2015,41(6):e74.
doi: 10.1093/nar/gkt006 |
| [42] | Zhao J, Wu J, Xu T, Yang Q, He J, Song X . IRESfinder: Identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J Genet Genomics, 2018,45(7):403-406. |
| [43] | Mokrejs M, Vopálenský V, Kolenaty O, Masek T, Feketová Z, Sekyrová P, Skaloudová B, Kríz V, Pospísek M . IRESite: the database of experimentally verified IRES structures. Nucleic Acids Res, 2006,34:125-130. |
| [44] | Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q . SRAMP: prediction of mammalian N 6-methyladenosine (m 6A) sites based on sequence-derived features . Nucleic Acids Res, 2016,44(10):e91. |
| [45] | Wei LY, Chen HR, Su R . M6APred-EL: A sequence-based predictor for identifying N 6-methyladenosine sites using ensemble learning . Mol Ther Nuceicl Acids, 2018,12:635-644. |
| [46] | Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer ELL, Tate J, Punta M . Pfam: the protein families database. Nucleic Acids Res, 2013,42(1):222-230. |
| [47] | Sun P, Li GL . CircCode: a powerful tool for identifying circRNA coding ability. Front Genet, 2019,10:981. |
| [48] | Chen XP, Han P, Zhou T, Guo XJ, Song XF, Li Y. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep, 2016,6:34985. |
| [49] | Liu M, Wang Q, Shen J, Yang BB, Ding XM . Circbank: a comprehensive database for circRNA with standard nomenclature. Rna Biol, 2019,7:899-905. |
| [50] | Glažar P, Papavasileiou P, Rajewsky N . CircBase: a database for circular RNAs. RNA, 2014,20(11):1666-1670. |
| [51] | Xia SY, Feng J, Chen K, Ma YB, Gong J, Cai FF, Jin YX, Gao Y, Xia LJ, Chang H, Wei L, Han L, He CJ . CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res, 2017,46(D1):925-929. |
| [52] | Xia SY, Feng J, Lei LJ, Hu J, Xia LJ, Wang J, Xiang Y, Liu LJ, Zhong S, Han L, He CJ . Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform, 2016,18(6):984-992. |
| [53] | Dong R, Ma XK, Li GW, Yang L . CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genom Proteom Bioinf, 2018,16(4):226-233. |
| [54] | Panda AC, Dudekula DB, Abdelmohsen K, Gorospe M . Analysis of circular RNAs using the web tool CircInteractome. Methods Mol Biol, 2018,1724:43-56. |
| [55] | Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin MF, Weng SL, Hsu SD, Huang CC, Cheng C, Liu CC, Huang HD . CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res, 2016,44(1):209-215. |
| [56] | Chu Q, Zhang X, Zhu X, Liu C, Mao L, Ye C, Zhu QH, Fan L . PlantcircBase: a database for plant circular RNAs. Mol Plant, 2017,10(8):1126-1128. |
| [57] | Ghosal S, Das S, Sen R, Basak P, Chakrabarti J . Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet, 2013,4:283. |
| [58] | Ji PF, Wu WY, Chen S, Zheng Y, Zhou L, Zhang JY, Cheng H, Yan J, Zhang SG, Yang PH, Zhao FQ. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep, 2019,26(12): 3444-3460. e5. |
| [59] | Ingolia NT, Lareau LF, Weissman JS . Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell, 2011,147(4):789-802. |
| [60] | Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS . Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science, 2009,324(5924):218-223. |
| [61] | Xia X, Li XX, Li FY, Wu XJ, Zhang ML, Zhou HK, Huang NN, Yang XS, Xiao FZ, Liu DW, Yang LX, Zhang N . A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer, 2019,18(1):131. |
| [62] | Tenenbaum SA, Lager PJ, Carson CC, Keene JD . Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods, 2002,26(2):191-198. |
| [63] | Marín-Béjar O, Huarte M . RNA pulldown protocol for in vitro detection and identification of RNA-associated proteins. Methods Mol Biol, 2015,1206:87-95. |
| [64] | Zhang ML, Zhao K, Xu XP, Yang YB, Yan S, Wei P, Liu H, Xu JB, Xiao FZ, Zhou HK, Yang XS, Huang NN, Liu JL, He KJ, Xie KP, Zhang G, Huang SY, Zhang N . A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun, 2018,9(1):4475. |
| [65] | Liang WC, Wong CW, Liang PP, Shi M, Cao Y, Rao ST, Tsui SKW, Waye MMY, Zhang Q, Fu WM, Zhang JF . Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol, 2019,20(1):84. |
| [66] | Gu CH, Zhou NC, Wang ZY, Li GR, Kou YP, Yu SL, Feng YJ, Chen L, Yang JJ, Tian FY . CircGprc5a promoted bladder oncogenesis and metastasis through Gprc5a- targeting peptide. Mol Ther Nucleic Acids, 2018,13:633-641. |
| [67] | Van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann CL, Kanda M, Worth CL, Schafer S, Calviello L, Merriott R, Patone G, Hummel O, Wyler E, Obermayer B, Mücke MB, Lindberg EL, Trnka F, Memczak S, Schilling M, Felkin LE, Barton PJR, Quaife NM, Vanezis K, Diecke S, Mukai M, Mah N, Oh S-J, Kurtz A, Schramm C, Schwinge D, Sebode M, Harakalova M, Asselbergs FW, Vink A, de Weger RA, Viswanathan S, Widjaja AA, Gärtner-Rommel A, Milting H, dos Remedios C, Knosalla C, Mertins P, Landthaler M, Vingron M, Linke WA, Seidman JG, Seidman CE, Rajewsky N, Ohler U, Cook SA, Hubner N. The translational landscape of the human heart. Cell, 2019,178(1):242-260. |
| [68] | Zhang ML, Huang NN, Yang XS, Luo JY, Yan S, Xiao FZ, Chen WP, Gao XY, Zhao K, Zhou HK, Li ZQ, Ming L, Xie B, Zhang N . A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene, 2018,37(13):1805-1814. |
| [69] | Wesselhoeft RA, Kowalski PS, Anderson DG . Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun, 2018,9(1):2629. |
| [70] | Wesselhoeft RA, Kowalski PS, Parker-Hale FC, Huang YX, Bisaria N, Anderson DG . RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol Cell, 2019,74(3):508-520. |
| [1] | 张为露, 冷奇颖, 郑嘉辉, AliHassanNawaz, 焦振海, 王府建, 张丽. 小鼠生长激素受体基因环状转录本的克隆及其表达规律[J]. 遗传, 2021, 43(9): 890-900. |
| [2] | 邢宝松, 王璟, 陈俊峰, 马强, 任巧玲, 张家庆, 张华, 滑留帅, 孙加节, 曹海. 去势和非去势公猪背最长肌circRNA差异表达分析[J]. 遗传, 2021, 43(11): 1066-1077. |
| [3] | 郑婷, 甘麦邻, 沈林園, 牛丽莉, 郭宗义, 王金勇, 张顺华, 朱砺. circRNA及其调控动物骨骼肌发育研究进展[J]. 遗传, 2020, 42(12): 1178-1191. |
| [4] | 刘旭庆,高宇帮,赵良真,蔡宇晨,王汇源,苗苗,顾连峰,张航晓. 环状RNA的产生、研究方法及功能[J]. 遗传, 2019, 41(6): 469-485. |
| [5] | 冷奇颖, 郑嘉辉, 徐海冬, PatriciaAdu-Asiamah, 张颖, 杜炳旺, 张丽. 鸡胰岛素降解酶基因环状转录本克隆及其表达规律[J]. 遗传, 2019, 41(12): 1129-1137. |
| [6] | 李静秋, 杨杰, 周平, 乐燕萍, 龚朝辉. 竞争性内源RNA的生物学功能及其调控[J]. 遗传, 2015, 37(8): 756-764. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||

www.chinagene.cn
备案号:京ICP备09063187号-4
总访问:,今日访问:,当前在线:
