Hereditas(Beijing) ›› 2021, Vol. 43 ›› Issue (1): 84-93.doi: 10.16288/j.yczz.20-337
• Research Article • Previous Articles Next Articles
Effect of Uhrf1 on intestinal development
Xinyue Wang1,3, Liang Li2,3, Qiuhui Duan2, Dali Li2( ), Jinlian Chen3(
), Jinlian Chen3( )
)
- 1. School of Medicine, Anhui University of Science and Technology, Huainan 232000, China
2. School of Life Sciences, East China Normal University, Shanghai Key Laboratory of Regulatory Biology, Shanghai 200241, China
3. Department of Gastroenterology, South Campus of Sixth People's Hospital, Shanghai Jiaotong University, Shanghai 201499, China
-
Received:2020-11-26Revised:2020-12-22Online:2021-01-20Published:2021-01-04 -
Contact:Li Dali,Chen Jinlian E-mail:dlli@bio.ecnu.edu.cn;wqq_021002@163.com -
Supported by:Supported by the National Natural Science Foundation of Shanghai No(16ZR1429100)
Cite this article
Xinyue Wang, Liang Li, Qiuhui Duan, Dali Li, Jinlian Chen. Effect of Uhrf1 on intestinal development[J]. Hereditas(Beijing), 2021, 43(1): 84-93.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
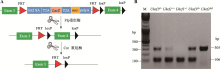
Table 1
The primer sequences of qRT-PCR"
| 基因 | 引物序列(5′→3′) |
|---|---|
| Lgr5 | F: CAGTGTTGTGCATTTGGGGG |
| R: CAAGGTCCCGCTCATCTTGA | |
| Sox9 | F: CACAAGAAAGACCACCCCGA |
| R: GGACCCTGAGATTGCCCAGA | |
| Ascl2 | F: CGTGAAGCTGGTGAACTTGG |
| R: GGATGTACTCCACGGCTGAG | |
| Olfm4 | F: TCTTGGGCAGAAGGTGGGACT |
| R: GGACCGTCAGGTTCAGGAGC | |
| Uhrf1 | F: ACGGTGCCTACTCATTGGTC |
| R: GCTTCTGGTCAGAGGACTGG | |
| β-actin | F: CAGCCTTCCTTCTTGGGTAT |
| R: TGATCTTGATCTTCATGGTGC |
| [1] |
Smith ZD, Meissner A . DNA methylation: roles in mammalian development. Nat Rev Genet, 2013,14(3):204-220.
doi: 10.1038/nrg3354 |
| [2] |
Greenberg MVC , Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol, 2019,20(10):590-607.
doi: 10.1038/s41580-019-0159-6 pmid: 31399642 |
| [3] |
Schübeler D . Function and information content of DNA methylation. Nature, 2015,517(7534):321-326.
doi: 10.1038/nature14192 pmid: 25592537 |
| [4] |
Smith ZD, Sindhu C, Meissner A . Molecular features of cellular reprogramming and development. Nat Rev Mol Cell Biol, 2016,17(3):139-154.
doi: 10.1038/nrm.2016.6 pmid: 26883001 |
| [5] |
Newkirk SJ, An WF . Uhrf1: a jack of all trades, and a master epigenetic regulator during spermatogenesis. Biol Reprod, 2020,102(6):1147-1152.
doi: 10.1093/biolre/ioaa026 pmid: 32101289 |
| [6] |
Xue BS, Zhao JS, Feng PH, Xing J, Wu HL, Li Y . Epigenetic mechanism and target therapy of Uhrf1 protein complex in malignancies. Onco Targets Ther, 2019,12:549-559.
doi: 10.2147/OTT.S192234 pmid: 30666134 |
| [7] |
Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H . The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature, 2007,450(7171):908-912.
doi: 10.1038/nature06397 pmid: 17994007 |
| [8] |
Xie S, Qian CM . The growing complexity of Uhrf1- mediated maintenance DNA methylation. Genes (Basel), 2018,9(12):600.
doi: 10.3390/genes9120600 |
| [9] |
Cheng JD, Yang Y, Fang J, Xiao JX, Zhu TT, Chen F, Wang P, Li Z, Yang HR, Xu YH . Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of Uhrf1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J Biol Chem, 2013,288(2):1329-1339.
doi: 10.1074/jbc.M112.415398 pmid: 23161542 |
| [10] |
Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, Koseki H, Nakanishi M . Uhrf1- dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature, 2013,502(7470):249-253.
doi: 10.1038/nature12488 |
| [11] |
Harrison JS, Cornett EM, Goldfarb D, DaRosa PA, Li ZM, Yan F, Dickson BM, Guo AH, Cantu DV, Kaustov L, Brown PJ, Arrowsmith CH, Erie DA, Major MB, Klevit RE, Krajewski K, Kuhlman B, Strahl BD, Rothbart SB. Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by Uhrf1. eLife, 2016,5:e17101.
doi: 10.7554/eLife.17101 pmid: 27595565 |
| [12] |
Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C . The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene, 2005, 24(49):7337-7345.
doi: 10.1038/sj.onc.1208878 pmid: 16007129 |
| [13] |
Tian YY, Paramasivam M, Ghosal G, Chen D, Shen X, Huang YL, Akhter S, Legerski R, Chen JJ, Seidman MM, Qin J, Li L . Uhrf1 contributes to DNA damage repair as a lesion recognition factor and nuclease scaffold. Cell Rep, 2015,10(12):1957-1966.
doi: 10.1016/j.celrep.2015.03.038 pmid: 25818288 |
| [14] |
Maenohara S, Unoki M, Toh H, Ohishi H, Sharif J, Koseki H, Sasaki H . Role of Uhrf1 in de novo DNA methylation in oocytes and maintenance methylation in preimplantation embryos. PLoS Genet, 2017,13(10):e1007042.
doi: 10.1371/journal.pgen.1007042 pmid: 28976982 |
| [15] |
Jacob V, Chernyavskaya Y, Chen XT, Tan PS, Kent B, Hoshida Y, Sadler KC . DNA hypomethylation induces a DNA replication-associated cell cycle arrest to block hepatic outgrowth in Uhrf1 mutant zebrafish embryos. Development, 2015,142(3):510-521.
doi: 10.1242/dev.115980 pmid: 25564650 |
| [16] |
Zhang YW, Chen YS, Ma R, Jiang YW, Liu J, Lin YT, Chen SQ, Xia MY, Zou F, Zhang JS, Pan T, Wang L, Wei L, Zhang H . Uhrf1 controls thymocyte fate decisions through the epigenetic regulation of EGR1 expression. J Immunol, 2020,204(12):3248-3261.
doi: 10.4049/jimmunol.1901471 pmid: 32358021 |
| [17] |
Ramesh V, Bayam E, Cernilogar FM, Bonapace IM, Schulze M, Riemenschneider MJ, Schotta G, Götz M . Loss of Uhrf1 in neural stem cells leads to activation of retroviral elements and delayed neurodegeneration. Genes Dev, 2016,30(19):2199-2212.
doi: 10.1101/gad.284992.116 pmid: 27798843 |
| [18] |
Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA . DNMT1 maintains progenitor function in self-renewing somatic tissue. Nature, 2010,463(7280):563-567.
doi: 10.1038/nature08683 pmid: 20081831 |
| [19] |
Yang XD, Han W, Liu F . DNA methylation in vertebrate embryogenesis. Hereditas (Beijing), 2012,34(9):1108-1113.
pmid: 23017451 |
|
杨晓丹, 韩威, 刘峰 . DNA甲基化与脊椎动物胚胎发育. 遗传, 2012,34(9):1108-1113.
pmid: 23017451 |
|
| [20] |
Bostick M, Kim JK, Estève PO, Clark A, Pradhan S, Jacobsen SE . Uhrf1 plays a role in maintaining DNA methylation in mammalian cells. Science, 2007,317(5845):1760-1764.
doi: 10.1126/science.1147939 pmid: 17673620 |
| [21] |
Blanchart A, Navis AC, Assaife-Lopes N, Usoskin D, Aranda S, Sontheimer J, Ernfors P . Uhrf1 licensed self-renewal of active adult neural stem cells. Stem Cells, 2018,36(11):1736-1751.
doi: 10.1002/stem.2889 pmid: 29999568 |
| [22] |
Obata Y, Furusawa Y, Endo TA, Sharif J, Takahashi D, Atarashi K, Nakayama M, Onawa S, Fujimura Y, Takahashi M, Ikawa T, Otsubo T, Kawamura YI, Dohi T, Tajima S, Masumoto H, Ohara O, Honda K, Hori S, Ohno H, Koseki H, Hase K . The epigenetic regulator Uhrf1 facilitates the proliferation and maturation of colonic regulatory T cells. Nat Immunol, 2014,15(6):571-579.
doi: 10.1038/ni.2886 |
| [23] |
Sadler KC, Krahn KN, Gaur NA, Ukomadu C . Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, Uhrf1. Proc Natl Acad Sci USA, 2007,104(5):1570-1575.
doi: 10.1073/pnas.0610774104 pmid: 17242348 |
| [24] |
Yamashita M, Inoue K, Saeki N, Ideta-Otsuka M, Yanagihara Y, Sawada Y, Sakakibara I, Lee J, Ichikawa K, Kamei Y, Iimura T, Igarashi K, Takada Y, Imai Y. Uhrf1 is indispensable for normal limb growth by regulating chondrocyte differentiation through specific gene expression. Development, 2018, 145(1): dev157412.
doi: 10.1242/dev.160051 pmid: 29217751 |
| [25] |
Jenkins Y, Markovtsov V, Lang W, Sharma P, Pearsall D, Warner J, Franci C, Huang B, Huang JN, Yam GC, Vistan JP, Pali E, Vialard J, Janicot M, Lorens JB, Payan DG, Hitoshi Y . Critical role of the ubiquitin ligase activity of Uhrf1, a nuclear RING finger protein, in tumor cell growth. Mol Biol Cell, 2005,16(12):5621-5629.
doi: 10.1091/mbc.e05-03-0194 pmid: 16195352 |
| [26] |
Ma HH, Chen H, Guo X, Wang ZT, Sowa ME, Zheng LJ, Hu SB, Zeng PY, Guo R, Diao JB, Lan F, Harper JW, Shi YG, Xu YH, Shi Y . M phase phosphorylation of the epigenetic regulator uhrf1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci USA, 2012,109(13):4828-4833.
doi: 10.1073/pnas.1116349109 pmid: 22411829 |
| [27] |
Loughery JEP, Dunne PD, O'Neill KM, Meehan RR, McDaid JR, Walsh CP.Dnmt1 deficiency triggers mismatch repair defects in human cells through depletion of repair protein levels in a process involving the DNA damage response. Hum Mol Genet, 2011, 20(16):3241-3255.
doi: 10.1093/hmg/ddr236 |
| [28] |
Tien AL, Senbanerjee S, Kulkarni A, Mudbhary R, Goudreau B, Ganesan S, Sadler KC, Ukomadu C . Uhrf1 depletion causes a G2/M arrest, activation of DNA damage response and apoptosis. Biochem J, 2011,435(1):175-185.
doi: 10.1042/BJ20100840 pmid: 21214517 |
| [29] |
Sheaffer KL, Kim R, Aoki R, Elliott EN, Schug J, Burger L, Schübeler D, Kaestner KH . DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev, 2014,28(6):652-664.
doi: 10.1101/gad.230318.113 pmid: 24637118 |
| [30] |
Elliott EN, Sheaffer KL, Schug J, Stappenbeck TS, Kaestner KH . Dnmt1 is essential to maintain progenitors in the perinatal intestinal epithelium. Development, 2015,142(12):2163-2172.
doi: 10.1242/dev.117341 pmid: 26023099 |
| [31] |
Elliott EN, Sheaffer KL, Kaestner KH . The 'de novo' DNA methyltransferase Dnmt3b compensates the Dnmt1-deficient intestinal epithelium. eLife, 2016,5:e12975.
doi: 10.7554/eLife.12975 pmid: 26808831 |
| [32] |
Chen C, Zhai SL, Zhang L, Chen JJ, Long XH, Qin J, Li JH, Huo R, Wang XM . Uhrf1 regulates germinal center B cell expansion and affinity maturation to control viral infection. J Exp Med, 2018,215(5):1437-1448.
doi: 10.1084/jem.20171815 pmid: 29618490 |
| [33] |
Xiang HD, Yuan LF, Gao X, Alexander PB, Lopez O, Lau C, Ding Y, Chong MY, Sun T, Chen R, Liu SQ, Wu HY, Wan Y, Randell SH, Li QJ, Wang XF . Uhrf1 is required for basal stem cell proliferation in response to airway injury. Cell Discov, 2017,3:17019.
doi: 10.1038/celldisc.2017.19 pmid: 28626588 |
| [34] |
Cui Y, Chen XF, Zhang JL, Sun X, Liu HF, Bai L, Xu CQ, Liu XL . Uhrf1 controls iNKT cell survival and differentiation through the Akt-mTOR axis. Cell Rep, 2016,15(2):256-263.
doi: 10.1016/j.celrep.2016.03.016 pmid: 27050515 |
| [1] | Mengxuan Xu, Ming Zhou. Advances of RNA polymerase IV in controlling DNA methylation and development in plants [J]. Hereditas(Beijing), 2022, 44(7): 567-580. |
| [2] | Wang Ya'nan, Tao Xu, Wanpeng Wang, Qingzhu Zhang, Xie Li'nan. Role of epigenetic modifications in the development of crops essential traits [J]. Hereditas(Beijing), 2021, 43(9): 858-879. |
| [3] | Xiangqian Zhang, Nan Li, Xinming Xie. Design and exploration of epigenetic comprehensive experiments [J]. Hereditas(Beijing), 2021, 43(12): 1179-1187. |
| [4] | Hengzhen Cui, Mizhu Sun, Runzhi Wang, Chenyu Li, Yuxuan Huang, Qiuju Huang, Xiaomeng Qiao. DNA methylation in the medial prefrontal cortex regulates alcohol-related behavior in rats [J]. Hereditas(Beijing), 2020, 42(1): 112-125. |
| [5] | Xinyuan Wang, Yu Zhang, Nan Yang, He Cheng, Yujie Sun. DNMT3a mediates paclitaxel-induced abnormal expression of LINE-1 by increasing the intragenic methylation [J]. Hereditas(Beijing), 2020, 42(1): 100-111. |
| [6] | Xin Huang,Yongqiang Chen,Guoliang Xu,Shuhong Peng. DNA methylation in adipose tissue and the development of diabetes and obesity [J]. Hereditas(Beijing), 2019, 41(2): 98-110. |
| [7] | Xuqiong Yang, Zhenfang Wu, Zicong Li. Advances in epigenetic reprogramming of somatic cells nuclear transfer in mammals [J]. Hereditas(Beijing), 2019, 41(12): 1099-1109. |
| [8] | Yunfeng Pan, Yanyi Wang, Jingwen Chen, Yimei Fan. Mitochondrial metabolism’s effect on epigenetic change and aging [J]. Hereditas(Beijing), 2019, 41(10): 893-904. |
| [9] | Junyi Ju,Quan Zhao. Regulation of γ-globin gene expression and its clinical applications [J]. Hereditas(Beijing), 2018, 40(6): 429-444. |
| [10] | Chendong Liu, Lu Yang, Hongzhou Pu, Qiong Yang, Wenyao Huang, Xue Zhao, Li Zhu, Shunhua Zhang. Epigenetics regulates gene expression patterns of skeletal muscle induced by physical exercise [J]. Hereditas(Beijing), 2017, 39(10): 888-896. |
| [11] | Ke Zhang, Guangde Feng, Baoyun Zhang, Wei Xiang, Long Chen, Fang Yang, Mingxing Chu, Pingqing Wang. Application of epigenetic markers in molecular breeding of the swine [J]. Hereditas(Beijing), 2016, 38(7): 634-643. |
| [12] | Yiran Zhu, Meiling Zhang, Zhichao Zhai, Yunjiao Zhao, Xin Ma. Epigenetic regulation of genomic imprinting in germline cells and preimplantation embryos [J]. HEREDITAS(Beijing), 2016, 38(2): 103-108. |
| [13] | Shuli Liu, Shengli Zhang, Ying Yu. Research progress of regulatory mechanism of DNA methylation in complex traits using monozygotic twins [J]. Hereditas(Beijing), 2016, 38(12): 1043-1055. |
| [14] | Yangyang Liu, Hengmi Cui. The method of estimating bisulfite conversion rate in DNA methylation analysis [J]. HEREDITAS(Beijing), 2015, 37(9): 939-944. |
| [15] | Longxiang Xie, Zhaoxiao Yu, Siyao Guo, Ping Li, Abualgasim Elgaili Abdalla, Jianping Xie. The roles of epigenetics and protein post-translational modifications in bacterial antibiotic resistance [J]. HEREDITAS(Beijing), 2015, 37(8): 793-800. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||