Hereditas(Beijing) ›› 2023, Vol. 45 ›› Issue (11): 950-962.doi: 10.16288/j.yczz.23-206
• Review • Previous Articles Next Articles
The application of CRISPR genome editing technologies in the pathogenesis studies, diagnosis, prevention and treatment of infectious diseases
Zhenrong Yang1,2( ), Gangqiao Zhou1,2(
), Gangqiao Zhou1,2( )
)
- 1. School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230022, China
2. State Key Lab of Proteomics, National Center for Protein Sciences at Beijing, Institute of Radiation Medicine, Academy of Military Medical Sciences, Academy of Military Sciences, Beijing 100850, China
-
Received:2023-07-31Revised:2023-10-06Online:2023-11-20Published:2023-10-24 -
Contact:Gangqiao Zhou E-mail:yangzr@mail.ustc.edu.cn;zhougq114@126.com
Cite this article
Zhenrong Yang, Gangqiao Zhou. The application of CRISPR genome editing technologies in the pathogenesis studies, diagnosis, prevention and treatment of infectious diseases[J]. Hereditas(Beijing), 2023, 45(11): 950-962.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Table 2
The application of CRISPR genome editing technologies in the diagnosis, prevention and treatment of infectious diseases"
| 应用类型 | 应用内容 | 实例 | 参考文献 |
|---|---|---|---|
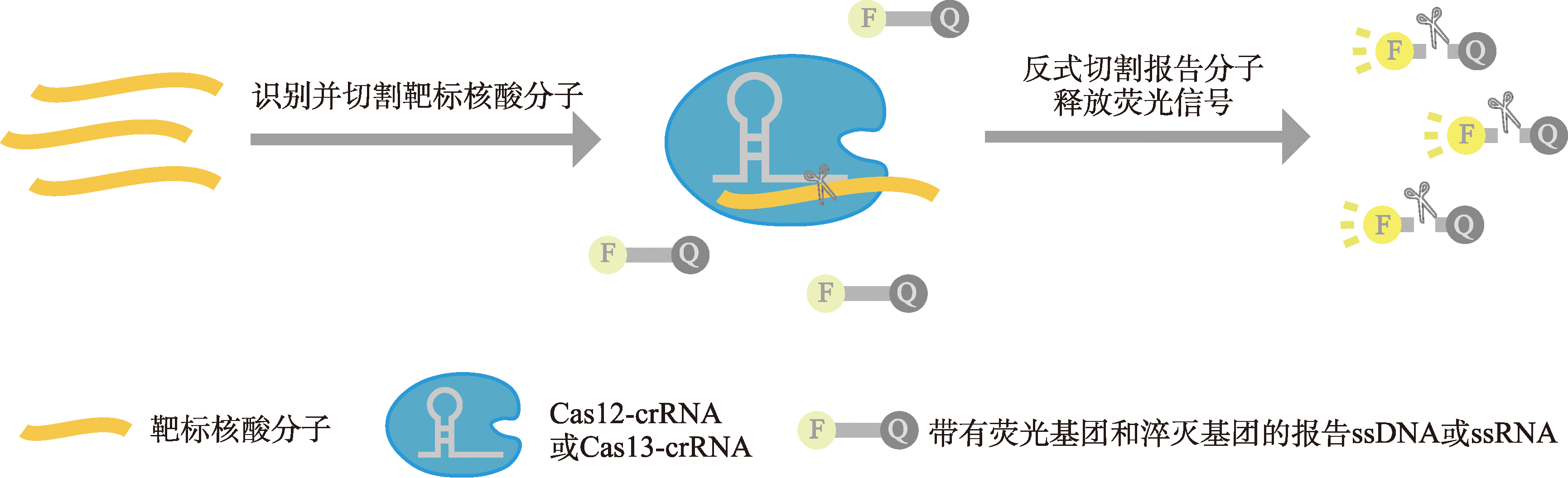
| 诊断 | 检测病原体核酸分子 | 利用基于CRISPR/Cas13a或CRISPR/Cas12b的核酸检测系统SHERLOCK/SHERLOCKv2检测ZIKV、DENV、WNV、YFV、SARS-CoV-2、大肠杆菌、铜绿假单胞菌和肺炎克雷伯菌; | [ |
| 利用基于CRISPR/Cas12a的DNA分子检测系统DETECTR检测HPV。 | [ | ||
| 预防 | 改造病原携带者基因组 | 利用CRISPR/Cas9工具将针对人类恶性疟原虫的单链抗体基因(m1C3,m2A10)敲入斯氏按蚊印度品系的胚胎中,获得携带有抗虫基因的后代; | [ |
| 利用CRISPR/Cas9工具靶向生育相关的基因(雌性冈比亚按蚊的dsxF基因,雄性埃及伊蚊的B2t基因),使突变蚊子不能产生后代。 | [ | ||
| 治疗 | 编辑细胞基因组/病毒基因组 | 利用CRISPR/Cas9工具编辑细胞的CCR5基因,或同时切除整合到细胞基因组上的HIV-1 DNA,以降低病毒载量; | [ |
| 利用CRISPR/Cas9工具切割受HBV感染细胞中的cccDNA,以减少总病毒DNA和cccDNA; | [ | ||
| 利用CRISPR/Cas9工具修饰受HSV-1感染细胞中的病毒基因组DNA,以减弱病毒毒力; | [ | ||
| 利用基于CRISPR/Cas13a的集诊断与抗病毒治疗于一体的CARVER系统检测并切割受LCMV、IAV或VSV感染细胞中的病毒基因组,以降低病毒RNA水平。 | [ |
| [1] |
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science, 2007, 315(5819): 1709-1712.
doi: 10.1126/science.1138140 pmid: 17379808 |
| [2] |
Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys, 2005, 38(1): 49-95.
pmid: 16336743 |
| [3] |
Bos JL, Heyting C, Borst P, Arnberg AC, Van Bruggen EF. An insert in the single gene for the large ribosomal RNA in yeast mitochondrial DNA. Nature, 1978, 275(5678): 336-338.
doi: 10.1038/275336a0 |
| [4] |
Stoddard BL. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob DNA, 2014, 5(1): 7.
doi: 10.1186/1759-8753-5-7 |
| [5] |
Sargent RG, Brenneman MA, Wilson JH. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol, 1997, 17(1): 267-277.
doi: 10.1128/MCB.17.1.267 pmid: 8972207 |
| [6] |
Cohen-Tannoudji M, Robine S, Choulika A, Pinto D, El Marjou F, Babinet C, Louvard D, Jaisser F. I-SceI-induced gene replacement at a natural locus in embryonic stem cells. Mol Cell Biol, 1998, 18(3): 1444-1448.
doi: 10.1128/MCB.18.3.1444 pmid: 9488460 |
| [7] | Cabaniols JP, Pâques F. Robust cell line development using meganucleases. Methods Mol Biol, 2008, 435: 31-45. |
| [8] |
Cabaniols JP, Ouvry C, Lamamy V, Fery I, Craplet ML, Moulharat N, Guenin SP, Bedut S, Nosjean O, Ferry G, Devavry S, Jacqmarcq C, Lebuhotel C, Mathis L, Delenda C, Boutin JA, Duchâteau P, Cogé F, Pâques F. Meganuclease-driven targeted integration in CHO-K1 cells for the fast generation of HTS-compatible cell-based assays. J Biomol Screen, 2010, 15(8): 956-967.
doi: 10.1177/1087057110375115 |
| [9] |
Redondo P, Prieto J, Muñoz IG, Alibés A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, Duchateau P, Pâques F, Blanco FJ, Montoya G. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature, 2008, 456(7218): 107-111.
doi: 10.1038/nature07343 |
| [10] |
Seligman LM, Stephens KM, Savage JH, Monnat RJ. Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics, 1997, 147(4): 1653-1664.
doi: 10.1093/genetics/147.4.1653 pmid: 9409828 |
| [11] |
Pósfai G, Kolisnychenko V, Bereczki Z, Blattner FR. Markerless gene replacement in Escherichia coli stimulated by a double-strand break in the chromosome. Nucleic Acids Res, 1999, 27(22): 4409-4415.
doi: 10.1093/nar/27.22.4409 pmid: 10536150 |
| [12] |
Puchta H, Dujon B, Hohn B. Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA, 1996, 93(10): 5055-5060.
doi: 10.1073/pnas.93.10.5055 pmid: 8643528 |
| [13] |
Antunes MS, Smith JJ, Jantz D, Medford JI. Targeted DNA excision in Arabidopsis by a re-engineered homing endonuclease. BMC Biotechnol, 2012, 12: 86.
doi: 10.1186/1472-6750-12-86 |
| [14] |
Djukanovic V, Smith J, Lowe K, Yang MZ, Gao HR, Jones S, Nicholson MG, West A, Lape J, Bidney D, Carl Falco S, Jantz D, Alexander Lyznik L. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P450-like gene (MS26) using a re-designed I-CreI homing endonuclease. Plant J, 2013, 76(5): 888-899.
doi: 10.1111/tpj.2013.76.issue-5 |
| [15] |
Rouet P, Smih F, Jasin M. Introduction of double- strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol, 1994, 14(12): 8096-8106.
doi: 10.1128/mcb.14.12.8096-8106.1994 pmid: 7969147 |
| [16] |
Gouble A, Smith J, Bruneau S, Perez C, Guyot V, Cabaniols JP, Leduc S, Fiette L, Avé P, Micheau B, Duchateau P, Pâques F. Efficient in toto targeted recombination in mouse liver by meganuclease-induced double-strand break. J Gene Med, 2006, 8(5): 616-622.
pmid: 16475243 |
| [17] |
Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA, 1996, 93(3): 1156-1160.
doi: 10.1073/pnas.93.3.1156 pmid: 8577732 |
| [18] |
Cathomen T, Keith Joung J,. Zinc-finger nucleases: the next generation emerges. Mol Ther, 2008, 16(7): 1200-1207.
doi: 10.1038/mt.2008.114 pmid: 18545224 |
| [19] |
Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 Å. Science, 1991, 252(5007): 809-817.
pmid: 2028256 |
| [20] |
Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc Natl Acad Sci USA, 1998, 95(18): 10570-10575.
doi: 10.1073/pnas.95.18.10570 pmid: 9724744 |
| [21] |
Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics, 2002, 161(3): 1169-1175.
doi: 10.1093/genetics/161.3.1169 pmid: 12136019 |
| [22] |
Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science, 2003, 300(5620): 764.
doi: 10.1126/science.1079512 pmid: 12730594 |
| [23] |
Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci USA, 2006, 103(44): 16370-16375.
doi: 10.1073/pnas.0605633103 pmid: 17060623 |
| [24] |
Meng XD, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol, 2008, 26(6): 695-701.
doi: 10.1038/nbt1398 pmid: 18500337 |
| [25] |
Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol, 2008, 26(6): 702-708.
doi: 10.1038/nbt1409 pmid: 18500334 |
| [26] |
Connelly JP, Barker JC, Pruett-Miller S, Porteus MH. Gene correction by homologous recombination with zinc finger nucleases in primary cells from a mouse model of a generic recessive genetic disease. Mol Ther, 2010, 18(6): 1103-1110.
doi: 10.1038/mt.2010.57 pmid: 20389291 |
| [27] |
Meyer M, de Angelis MH, Wurst W, Kühn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA, 2010, 107(34): 15022-15026.
doi: 10.1073/pnas.1009424107 pmid: 20686113 |
| [28] |
Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, Cui XX. Targeted genome modification in mice using zinc-finger nucleases. Genetics, 2010, 186(2): 451-459.
doi: 10.1534/genetics.110.117002 pmid: 20628038 |
| [29] |
Cui XX, Ji D, Fisher DA, Wu YM, Briner DM, Weinstein EJ. Targeted integration in rat and mouse embryos with zinc-finger nucleases. Nat Biotechnol, 2011, 29(1): 64-67.
doi: 10.1038/nbt.1731 pmid: 21151125 |
| [30] |
Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med, 2014, 370(10): 901-910.
doi: 10.1056/NEJMoa1300662 |
| [31] |
Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics, 2010, 186(2): 757-761.
doi: 10.1534/genetics.110.120717 pmid: 20660643 |
| [32] |
Bak RO, Gomez-Ospina N, Porteus MH. Gene editing on center stage. Trends Genet, 2018, 34(8): 600-611.
doi: S0168-9525(18)30089-1 pmid: 29908711 |
| [33] | Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol, 2013, 14(1): 49-55. |
| [34] |
Qasim W, Zhan H, Samarasinghe S, Adams S, Amrolia P, Stafford S, Butler K, Rivat C, Wright G, Somana K, Ghorashian S, Pinner D, Ahsan G, Gilmour K, Lucchini G, Inglott S, Mifsud W, Chiesa R, Peggs KS, Chan L, Farzeneh F, Thrasher AJ, Vora A, Pule M, Veys P. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med, 2017, 9(374): eaaj2013.
doi: 10.1126/scitranslmed.aaj2013 |
| [35] |
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol, 1987, 169(12): 5429-5433.
doi: 10.1128/jb.169.12.5429-5433.1987 pmid: 3316184 |
| [36] |
Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol, 2000, 36(1): 244-246.
doi: 10.1046/j.1365-2958.2000.01838.x pmid: 10760181 |
| [37] |
Jansen R, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol, 2002, 43(6): 1565-1575.
doi: 10.1046/j.1365-2958.2002.02839.x pmid: 11952905 |
| [38] |
Wang JY, Doudna JA. CRISPR technology: a decade of genome editing is only the beginning. Science, 2023, 379(6629): eadd8643.
doi: 10.1126/science.add8643 |
| [39] |
Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol, 2005, 60(2): 174-182.
doi: 10.1007/s00239-004-0046-3 pmid: 15791728 |
| [40] |
Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology (Reading), 2005, 151(Pt 3): 653-663.
doi: 10.1099/mic.0.27437-0 |
| [41] |
Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading), 2005, 151(Pt 8): 2551-2561.
doi: 10.1099/mic.0.28048-0 |
| [42] |
Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct, 2006, 1: 7.
pmid: 16545108 |
| [43] |
Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas Č, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, van der Oost J, Barrangou R, Koonin EV. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol, 2020, 18(2): 67-83.
doi: 10.1038/s41579-019-0299-x pmid: 31857715 |
| [44] |
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 2012, 337(6096): 816-821.
doi: 10.1126/science.1225829 pmid: 22745249 |
| [45] |
Doudna JA, Charpentier E.The new frontier of genome engineering with CRISPR-Cas9. Science, 2014, 346(6213): 1258096.
doi: 10.1126/science.1258096 |
| [46] |
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F.Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 2015, 163(3): 759-771.
doi: 10.1016/j.cell.2015.09.038 pmid: 26422227 |
| [47] |
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DBT, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science, 2016, 353(6299): aaf5573.
doi: 10.1126/science.aaf5573 |
| [48] | Chen JS, Ma EB, Harrington LB, Da Costa M, Tian XR, Palefsky JM, Doudna JA. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, 2018, 360(6387): 436-439. |
| [49] |
Broughton JP, Deng XD, Yu GX, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, Zorn K, Gopez A, Hsu E, Gu W, Miller S, Pan CY, Guevara H, Wadford DA, Chen JS, Chiu CY.CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol, 2020, 38(7): 870-874.
doi: 10.1038/s41587-020-0513-4 pmid: 32300245 |
| [50] |
Nguyen LT, Smith BM, Jain PK. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat Commun, 2020, 11(1): 4906.
doi: 10.1038/s41467-020-18615-1 pmid: 32999292 |
| [51] |
Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Myhrvold C, Bhattacharyya RP, Livny J, Regev A, Koonin EV, Hung DT, Sabeti PC, Collins JJ, Zhang F.Nucleic acid detection with CRISPR-Cas13a/C2c2. Science, 2017, 356(6336): 438-442.
doi: 10.1126/science.aam9321 pmid: 28408723 |
| [52] |
Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F.Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science, 2018, 360(6387): 439-444.
doi: 10.1126/science.aaq0179 pmid: 29449508 |
| [53] |
Iwasaki RS, Batey RT. SPRINT: a Cas13a-based platform for detection of small molecules. Nucleic Acids Res, 2020, 48(17): e101.
doi: 10.1093/nar/gkaa673 |
| [54] | Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA, 2012, 109(39): E2579-E2586. |
| [55] |
Rees HA, Liu DR. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet, 2018, 19(12): 770-788.
doi: 10.1038/s41576-018-0059-1 |
| [56] |
Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR- Cas9 complex. Nature, 2015, 517(7536): 583-588.
doi: 10.1038/nature14136 |
| [57] |
Gilbert LA, Larson MH, Morsut L, Liu ZR, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR- mediated modular RNA-guided regulation of transcription in eukaryotes. Cell, 2013, 154(2): 442-451.
doi: 10.1016/j.cell.2013.06.044 pmid: 23849981 |
| [58] |
Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol, 2015, 33(5): 510-517.
doi: 10.1038/nbt.3199 pmid: 25849900 |
| [59] |
Zhang HX, Zhang Y, Yin H. Genome editing with mRNA encoding ZFN, TALEN, and Cas9. Mol Ther, 2019, 27(4): 735-746.
doi: 10.1016/j.ymthe.2019.01.014 |
| [60] |
Ren YX, Xiao RD, Lou XM, Fang XD. Research advance and application in the gene therapy of gene editing technologies. Hereditas(Beijing), 2019, 41(1): 18-27.
doi: 10.16288/j.yczz.18-142 pmid: 30686782 |
|
任云晓, 肖茹丹, 娄晓敏, 方向东. 基因编辑技术及其在基因治疗中的应用. 遗传, 2019, 41(1): 18-27.
doi: 10.16288/j.yczz.18-142 pmid: 30686782 |
|
| [61] |
McCarty NS, Graham AE, Studena L, Ledesma-Amaro R. Multiplexed CRISPR technologies for gene editing and transcriptional regulation. Nat Commun, 2020, 11(1): 1281.
doi: 10.1038/s41467-020-15053-x pmid: 32152313 |
| [62] |
Ye L, Wang JM, Beyer AI, Teque F, Cradick TJ, Qi ZX, Chang JC, Bao G, Muench MO, Yu JW, Levy JA, Kan YW. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA, 2014, 111(26): 9591-9596.
doi: 10.1073/pnas.1407473111 pmid: 24927590 |
| [63] |
Yusa K, Zhou LQ, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci USA, 2011, 108(4): 1531-1536.
doi: 10.1073/pnas.1008322108 pmid: 21205896 |
| [64] |
Zhou J, Li C, Liu XJ, Chiu MC, Zhao XY, Wang D, Wei YX, Lee A, Zhang AJ, Chu H, Cai JP, Yip CCY, Chan IHY, Wong KKY, Tsang OTY, Chan KH, Chan JFW, To KKW, Chen HL, Yuen KY.Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med, 2020, 26(7): 1077-1083.
doi: 10.1038/s41591-020-0912-6 pmid: 32405028 |
| [65] |
Lamers MM, Haagmans BL. SARS-CoV-2 pathogenesis. Nat Rev Microbiol, 2022, 20(5): 270-284.
doi: 10.1038/s41579-022-00713-0 pmid: 35354968 |
| [66] |
Sun SH, Chen Q, Gu HJ, Yang G, Wang YX, Huang XY, Liu SS, Zhang NN, Li XF, Xiong R, Guo Y, Deng YQ, Huang WJ, Liu Q, Liu QM, Shen YL, Zhou Y, Yang X, Zhao TY, Fan CF, Zhou YS, Qin CF, Wang YC. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe, 2020, 28(1): 124-133.e4.
doi: 10.1016/j.chom.2020.05.020 |
| [67] |
Li B, Clohisey SM, Chia BS, Wang B, Cui A, Eisenhaure T, Schweitzer LD, Hoover P, Parkinson NJ, Nachshon A, Smith N, Regan T, Farr D, Gutmann MU, Bukhari SI, Law A, Sangesland M, Gat-Viks I, Digard P, Vasudevan S, Lingwood D, Dockrell DH, Doench JG, Baillie JK, Hacohen N. Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat Commun, 2020, 11(1): 164.
doi: 10.1038/s41467-019-13965-x pmid: 31919360 |
| [68] |
Hyrina A, Jones C, Chen D, Clarkson S, Cochran N, Feucht P, Hoffman G, Lindeman A, Russ C, Sigoillot F, Tsang T, Uehara K, Xie LL, Ganem D, Holdorf M. A genome-wide CRISPR screen identifies ZCCHC14 as a host factor required for hepatitis B surface antigen production. Cell Rep, 2019, 29(10): 2970-2978.e6.
doi: S2211-1247(19)31446-9 pmid: 31801065 |
| [69] |
Mei H, Zha Z, Wang W, Xie YS, Huang YG, Li WP, Wei D, Zhang XX, Qu JM, Liu J. Surfaceome CRISPR screen identifies OLFML3 as a rhinovirus-inducible IFN antagonist. Genome Biol, 2021, 22(1): 297.
doi: 10.1186/s13059-021-02513-w pmid: 34686207 |
| [70] |
Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, Strine MS, Zhang SM, Graziano VR, Schmitz CO, Chen JS, Mankowski MC, Filler RB, Ravindra NG, Gasque V, de Miguel FJ, Patil A, Chen HC, Oguntuyo KY, Abriola L, Surovtseva YV, Orchard RC, Lee B, Lindenbach BD, Politi K, van Dijk D, Kadoch C, Simon MD, Yan Q, Doench JG, Wilen CB. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell, 2021, 184(1): 76-91.e13.
doi: 10.1016/j.cell.2020.10.028 pmid: 33147444 |
| [71] | Baggen J, Persoons L, Vanstreels E, Jansen S, Van Looveren D, Boeckx B, Geudens V, De Man J, Jochmans D, Wauters J, Wauters E, Vanaudenaerde BM, Lambrechts D, Neyts J, Dallmeier K, Thibaut HJ, Jacquemyn M, Maes P, Daelemans D.Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat Genet, 2021, 53(4): 435-444. |
| [72] |
Biering SB, Sarnik SA, Wang E, Zengel JR, Leist SR, Schäfer A, Sathyan V, Hawkins P, Okuda K, Tau C, Jangid AR, Duffy CV, Wei J, Gilmore RC, Alfajaro MM, Strine MS, Nguyenla X, Van Dis E, Catamura C, Yamashiro LH, Belk JA, Begeman A, Stark JC, Shon DJ, Fox DM, Ezzatpour S, Huang E, Olegario N, Rustagi A, Volmer AS, Livraghi-Butrico A, Wehri E, Behringer RR, Cheon DJ, Schaletzky J, Aguilar HC, Puschnik AS, Button B, Pinsky BA, Blish CA, Baric RS, O’Neal WK, Bertozzi CR, Wilen CB, Boucher RC, Carette JE, Stanley SA, Harris E, Konermann S, Hsu PD. Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection. Nat Genet, 2022, 54(8): 1078-1089.
doi: 10.1038/s41588-022-01131-x |
| [73] |
Zhu SY, Liu Y, Zhou Z, Zhang ZY, Xiao X, Liu ZH, Chen A, Dong XJ, Tian F, Chen SH, Xu YY, Wang CH, Li QH, Niu XR, Pan Q, Du S, Xiao JY, Wang JW, Wei WS. Genome-wide CRISPR activation screen identifies candidate receptors for SARS-CoV-2 entry. Sci China Life Sci, 2022, 65(4): 701-717.
doi: 10.1007/s11427-021-1990-5 |
| [74] | Lai Y, Cui L, Babunovic GH, Fortune SM, Doench JG, Lu TK. High-throughput CRISPR screens to dissect macrophage-Shigella interactions. mBio, 2021, 12(6): e0215821. |
| [75] | Bosch B, DeJesus MA, Poulton NC, Zhang WZ, Engelhart CA, Zaveri A, Lavalette S, Ruecker N, Trujillo C, Wallach JB, Li SQ, Ehrt S, Chait BT, Schnappinger D, Rock JM. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell, 2021, 184(17): 4579-4592.e24. |
| [76] |
Rousset F, Bikard D. CRISPR screens in the era of microbiomes. Curr Opin Microbiol, 2020, 57: 70-77.
doi: S1369-5274(20)30090-4 pmid: 32858412 |
| [77] | Yan MY, Zheng DD, Li SS, Ding XY, Wang CL, Guo XP, Zhan LJ, Jin Q, Yang J, Sun YC. Application of combined CRISPR screening for genetic and chemical-genetic interaction profiling in Mycobacterium tuberculosis. Sci Adv, 2022, 8(47): eadd5907. |
| [78] | Yan MY, Li SS, Ding XY, Guo XP, Jin Q, Sun YC. A CRISPR-assisted nonhomologous end-joining strategy for efficient genome editing in Mycobacterium tuberculosis. mBio, 2020, 11(1): e02364-19. |
| [79] | Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, Kellner MJ, Tan AL, Paul LM, Parham LA, Garcia KF, Barnes KG, Chak B, Mondini A, Nogueira ML, Isern S, Michael SF, Lorenzana I, Yozwiak NL, MacInnis BL, Bosch I, Gehrke L, Zhang F, Sabeti PC.Field-deployable viral diagnostics using CRISPR-Cas13. Science, 2018, 360(6387): 444-448. |
| [80] |
An JH, Liao XJ, Xiao TY, Qian S, Yuan J, Ye HC, Qi FR, Shen CG, Wang LF, Liu Y, Cheng XY, Li N, Cai QX, Wang F, Chen J, Li GJ, Cai QE, Liu YX, Wang YF, Zhang F, Fu Y, He Q, Tan XH, Liu L, Zhang Z. Clinical characteristics of recovered COVID-19 patients with re-detectable positive RNA test. Ann Transl Med, 2020, 8(17): 1084.
doi: 10.21037/atm-20-5602 pmid: 33145303 |
| [81] | Joung J, Ladha A, Saito M, Segel M, Bruneau R, Huang MLW, Kim NG, Yu X, Li J, Walker BD, Greninger AL, Jerome KR, Gootenberg JS, Abudayyeh OO, Zhang F. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv, 2020, doi: 10.1101/2020.05.04.20091231. |
| [82] |
Joung J, Ladha A, Saito M, Kim NG, Woolley AE, Segel M, Barretto RPJ, Ranu A, Macrae RK, Faure G, Ioannidi EI, Krajeski RN, Bruneau R, Huang MLW, Yu XG, Li JZ, Walker BD, Hung DT, Greninger AL, Jerome KR, Gootenberg JS, Abudayyeh OO, Zhang F. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med, 2020, 383(15): 1492-1494.
doi: 10.1056/NEJMc2026172 |
| [83] |
Ranson H, Lissenden N. Insecticide resistance in African Anopheles mosquitoes: a worsening situation that needs urgent action to maintain malaria control. Trends Parasitol, 2016, 32(3): 187-196.
doi: 10.1016/j.pt.2015.11.010 |
| [84] | Moyes CL, Vontas J, Martins AJ, Ng LC, Koou SY, Dusfour I, Raghavendra K, Pinto J, Corbel V, David JP, Weetman D. Contemporary status of insecticide resistance in the major Aedes vectors of arboviruses infecting humans. PLoS Negl Trop Dis, 2017, 11(7): e0005625. |
| [85] | Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA.Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci USA, 2015, 112(49): E6736-E6743. |
| [86] |
Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, Beaghton AK, Nolan T, Crisanti A. A CRISPR-Cas 9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat Biotechnol, 2018, 36(11): 1062-1066.
doi: 10.1038/nbt.4245 |
| [87] | Chen JY, Luo JJ, Wang YJ, Gurav AS, Li M, Akbari OS, Montell C. Suppression of female fertility in Aedes aegypti with a CRISPR-targeted male-sterile mutation. Proc Natl Acad Sci USA, 2021, 118(22): e2105075118. |
| [88] |
Xu L, Wang J, Liu YL, Xie LF, Su B, Mou DL, Wang LT, Liu TT, Wang XB, Zhang B, Zhao L, Hu LD, Ning HM, Zhang YF, Deng K, Liu LF, Lu XF, Zhang T, Xu J, Li C, Wu H, Deng HK, Chen H. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N Engl J Med, 2019, 381(13): 1240-1247.
doi: 10.1056/NEJMoa1817426 |
| [89] | Dash PK, Chen C, Kaminski R, Su H, Mancuso P, Sillman B, Zhang C, Liao SR, Sravanam S, Liu H, Waight E, Guo LL, Mathews S, Sariyer R, Mosley RL, Poluektova LY, Caocci M, Amini S, Gorantla S, Burdo TH, Edagwa B, Gendelman HE, Khalili K. CRISPR editing of CCR5 and HIV-1 facilitates viral elimination in antiretroviral drug-suppressed virus-infected humanized mice. Proc Natl Acad Sci USA, 2023, 120(19): e2217887120. |
| [90] |
Kennedy EM, Bassit LC, Mueller H, Kornepati AVR, Bogerd HP, Nie T, Chatterjee P, Javanbakht H, Schinazi RF, Cullen BR. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology, 2015, 476: 196-205.
doi: S0042-6822(14)00537-6 pmid: 25553515 |
| [91] |
Ramanan V, Shlomai A, Cox DBT, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep, 2015, 5: 10833.
doi: 10.1038/srep10833 pmid: 26035283 |
| [92] | Li H, Sheng CY, Wang S, Yang L, Liang Y, Huang Y, Liu HB, Li P, Yang CJ, Yang XX, Jia LL, Xie J, Wang LG, Hao RZ, Du XY, Xu DP, Zhou JJ, Li MZ, Sun YS, Tong YG, Li Q, Qiu SF, Song HB.Removal of integrated hepatitis B virus DNA using CRISPR-Cas9. Front Cell Infect Microbiol, 2017, 7: 91. |
| [93] |
Russell TA, Stefanovic T, Tscharke DC. Engineering herpes simplex viruses by infection-transfection methods including recombination site targeting by CRISPR/Cas9 nucleases. J Virol Methods, 2015, 213: 18-25.
doi: 10.1016/j.jviromet.2014.11.009 pmid: 25479355 |
| [94] |
Lin CL, Li HH, Hao MR, Xiong D, Luo Y, Huang CH, Yuan Q, Zhang J, Xia NS. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing of HSV-1 virus in human cells. Sci Rep, 2016, 6: 34531.
doi: 10.1038/srep34531 pmid: 27713537 |
| [95] |
Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS, Yozwiak NL, Zhang F, Sabeti PC.Programmable inhibition and detection of RNA viruses using Cas13. Mol Cell, 2019, 76(5): 826-837.e11.
doi: S1097-2765(19)30698-7 pmid: 31607545 |
| [96] |
Wilbie D, Walther J, Mastrobattista E. Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc Chem Res, 2019, 52(6): 1555-1564.
doi: 10.1021/acs.accounts.9b00106 |
| [97] | Niu XR, Yin SM, Chen X, Shao TT, Li DL. Gene editing technology and its recent progress in disease therapy. Hereditas(Beijing), 2019, 41(7): 582-598. |
| 牛煦然, 尹树明, 陈曦, 邵婷婷, 李大力. 基因编辑技术及其在疾病治疗中的研究进展. 遗传, 2019, 41(7): 582-598. |
| [1] | Yi Zhang, Zhi-Ying Wu. Pathogenesis and therapeutic advances of cerebral autosomal- dominant arteriopathy with subcortical infarcts and leukoencephalopathy [J]. Hereditas(Beijing), 2023, 45(7): 568-579. |
| [2] | Bingzheng Wang, Chao Zhang, Jiali Zhang, Jin Sun. Conditional editing of the Drosophila melanogaster genome using single transcripts expressing Cas9 and sgRNA [J]. Hereditas(Beijing), 2023, 45(7): 593-601. |
| [3] | Danni Liu, Haiping Wu, Guohua Zhou. Research progress of visual detection in rapid on-site detection of pathogen nucleic acid [J]. Hereditas(Beijing), 2023, 45(4): 306-323. |
| [4] | Zhongsheng Wu, Yu Gao, Yongtao Du, Song Dang, Kangmin He. The protocol of tagging endogenous proteins with fluorescent tags using CRISPR-Cas9 genome editing [J]. Hereditas(Beijing), 2023, 45(2): 165-175. |
| [5] | Beiping Zeng, Hongen Xu, Lu Mao, Wenxue Tang. Molecular diagnosis of hereditary deafness and application of stepwise testing strategy [J]. Hereditas(Beijing), 2023, 45(1): 29-41. |
| [6] | Meizhen Liu, Liren Wang, Yongmei Li, Xueyun Ma, Honghui Han, Dali Li. Generation of genetically modified rat models via the CRISPR/Cas9 technology [J]. Hereditas(Beijing), 2023, 45(1): 78-87. |
| [7] | Xiaojun Zhang, Kun Xu, Juncen Shen, Lu Mu, Hongrun Qian, Jieyu Cui, Baoxia Ma, Zhilong Chen, Zhiying Zhang, Zehui Wei. A CRISPR/Cas9-Gal4BD donor adapting system for enhancing homology-directed repair [J]. Hereditas(Beijing), 2022, 44(8): 708-719. |
| [8] | Chong Zhang, Zixuan Wei, Min Wang, Yaosheng Chen, Zuyong He. Editing MC1R in human melanoma cells by CRISPR/Cas9 and functional analysis [J]. Hereditas(Beijing), 2022, 44(7): 581-590. |
| [9] | Zhuoyuan Jiang, Yan Zha, Xiaofeng Shi, Yongbiao Zhang. Research progress on neural crest cells and neurocristopathies and its pathogenesis [J]. Hereditas(Beijing), 2022, 44(2): 117-133. |
| [10] | Yangjinghui Zhang, Peiyao Chang, Zishu Yang, Yuhang Xue, Xueqi Li, Yang Zhang. Advances in epigenetic modification affecting anthocyanin synthesis [J]. Hereditas(Beijing), 2022, 44(12): 1117-1127. |
| [11] | Min Shen, Yong Gu, Changjiang Ying, Mei Zhang, Tao Yang, Yang Chen. Diagnosis, treatment and genetic analysis of a case with fibrocalculous pancreatic diabetes [J]. Hereditas(Beijing), 2022, 44(11): 1079-1086. |
| [12] | Yao Liu, Xianhui Zhou, Shuhong Huang, Xiaolong Wang. Prime editing: a search and replace tool with versatile base changes [J]. Hereditas(Beijing), 2022, 44(11): 993-1008. |
| [13] | Jiaqi Liang, Chang Liu, Wenxiang Zhang, Siyu Chen. Interaction between hepatokines and metabolic diseases [J]. Hereditas(Beijing), 2022, 44(10): 853-866. |
| [14] | Yuting Han, Bowen Xu, Yutong Li, Xinyi Lu, Xizhi Dong, Yuhao Qiu, Qinyun Che, Ruibao Zhu, Li Zheng, Xiaochen Li, Xu Si, Jianquan Ni. The cutting edge of gene regulation approaches in model organism Drosophila [J]. Hereditas(Beijing), 2022, 44(1): 3-14. |
| [15] | Guangwu Yang, Yuan Tian. The F-box gene Ppa promotes lipid storage in Drosophila [J]. Hereditas(Beijing), 2021, 43(6): 615-622. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||