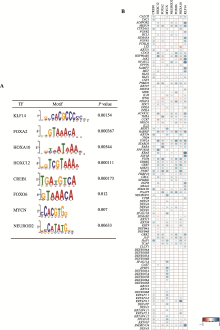
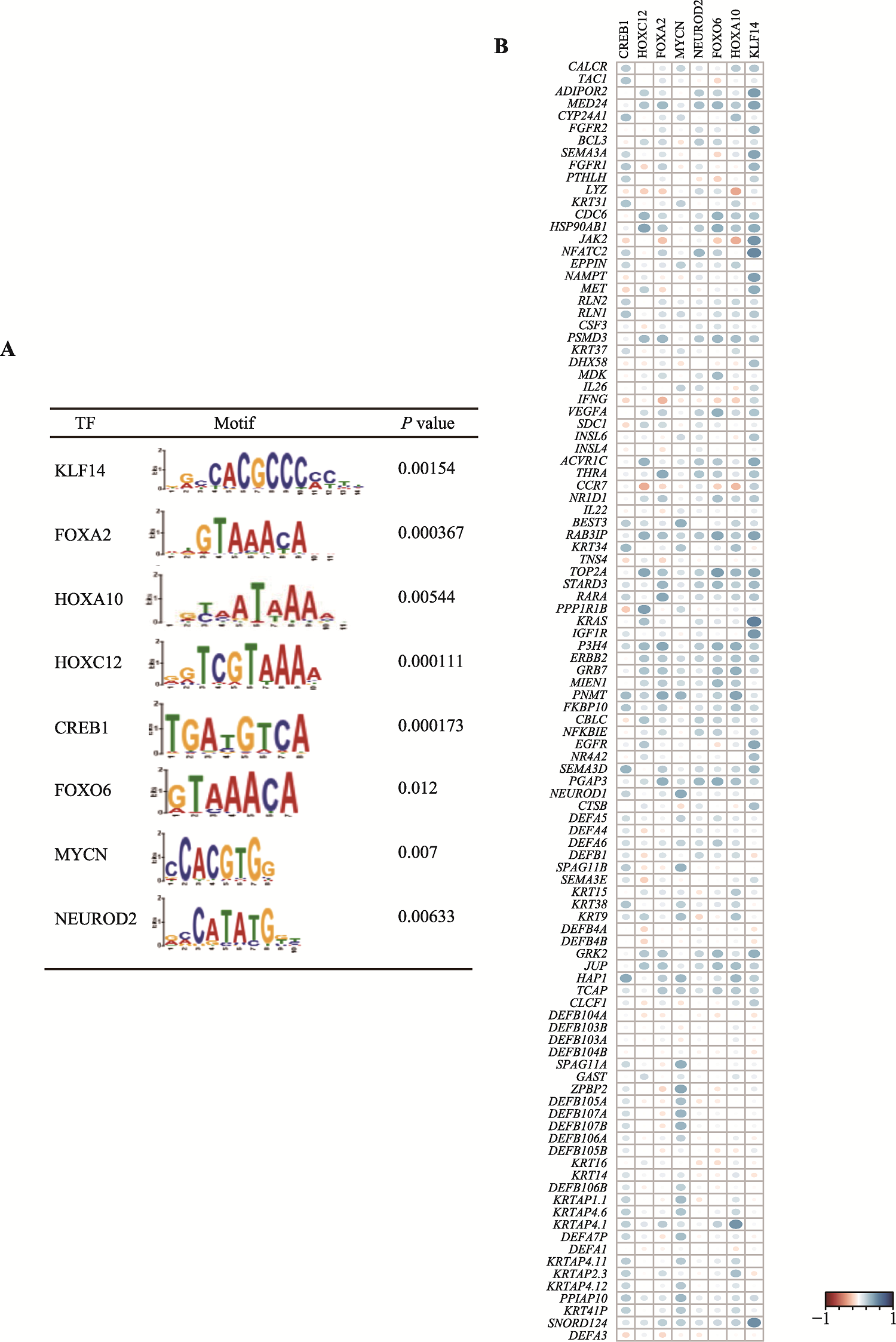
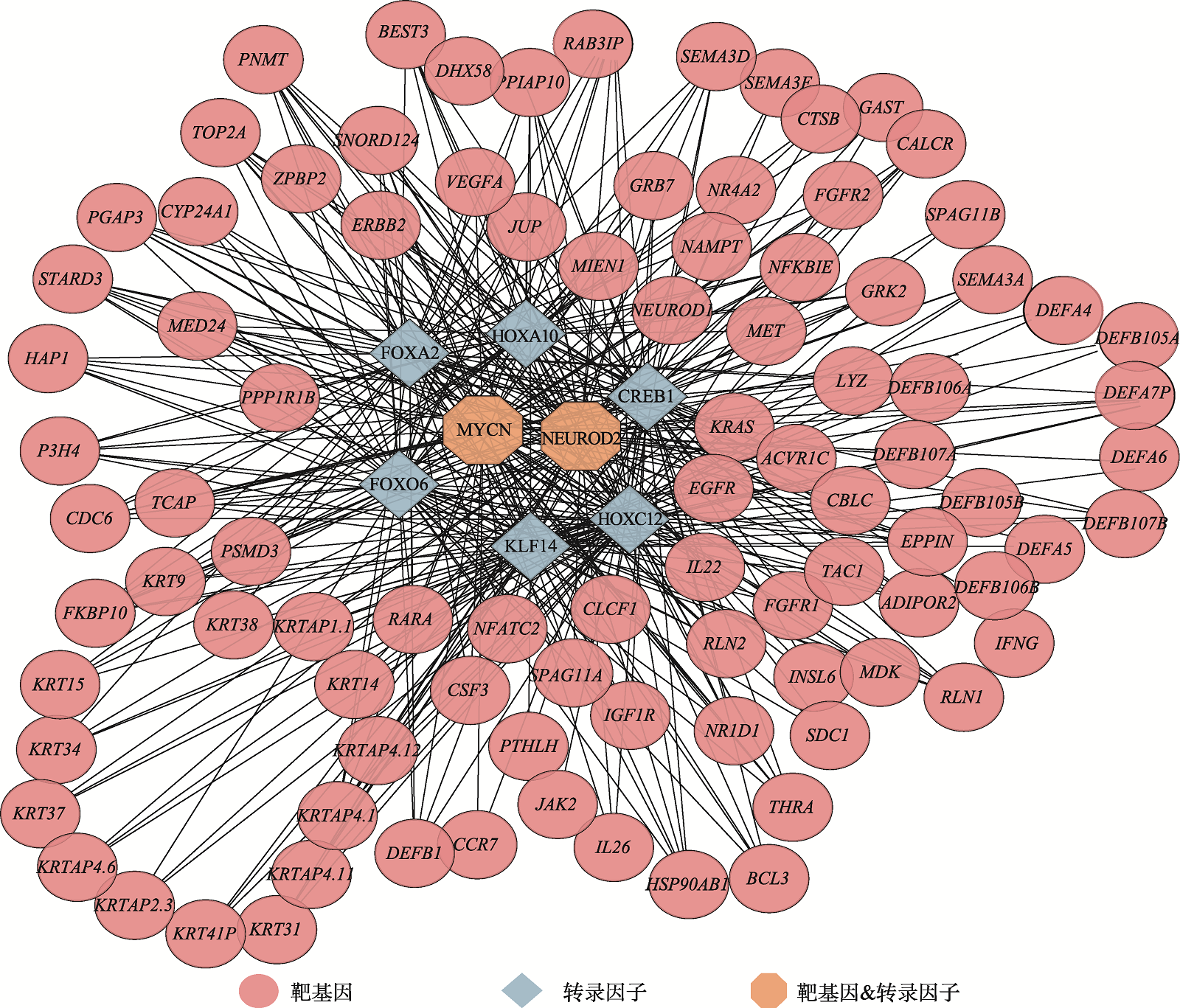
Hereditas(Beijing) ›› 2025, Vol. 47 ›› Issue (5): 558-572.doi: 10.16288/j.yczz.24-291
• Research Article • Previous Articles Next Articles
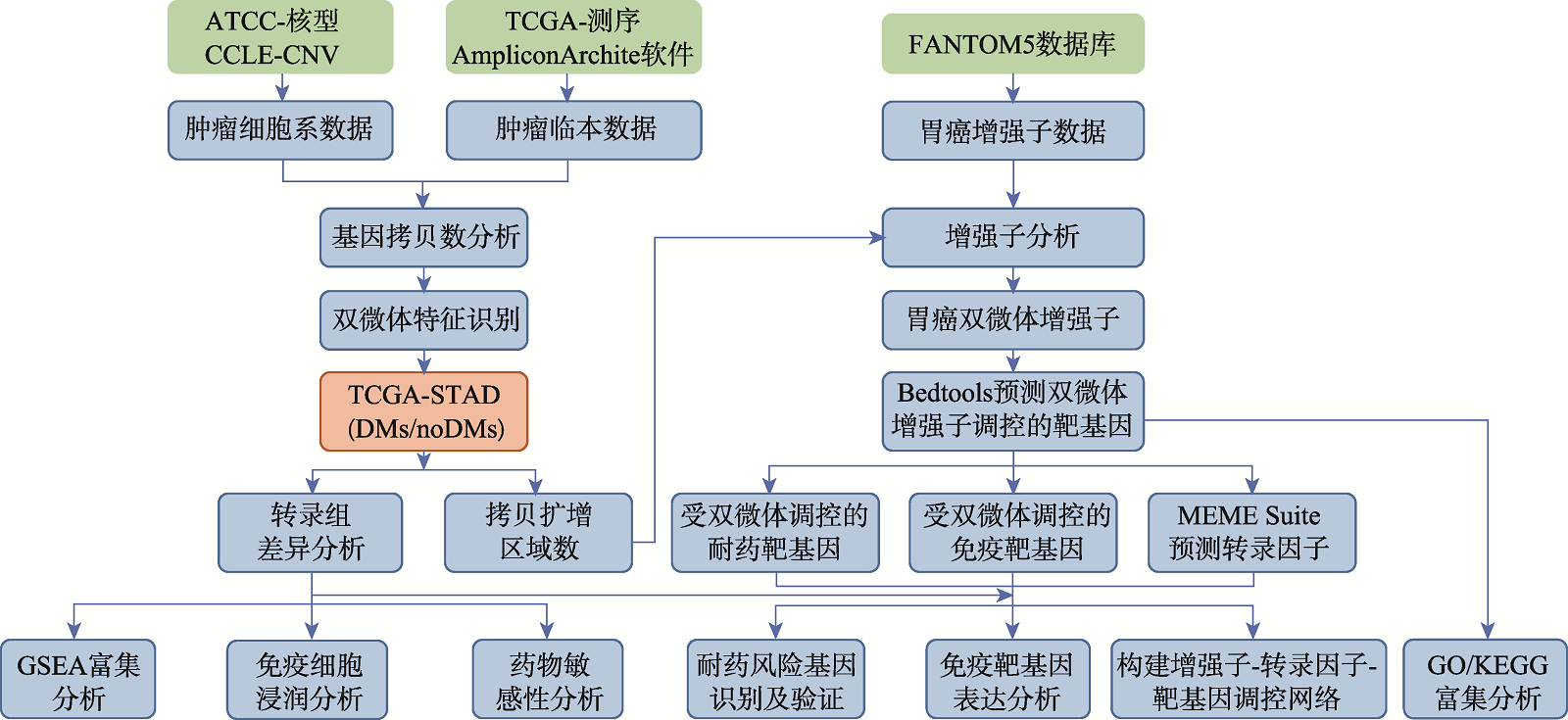
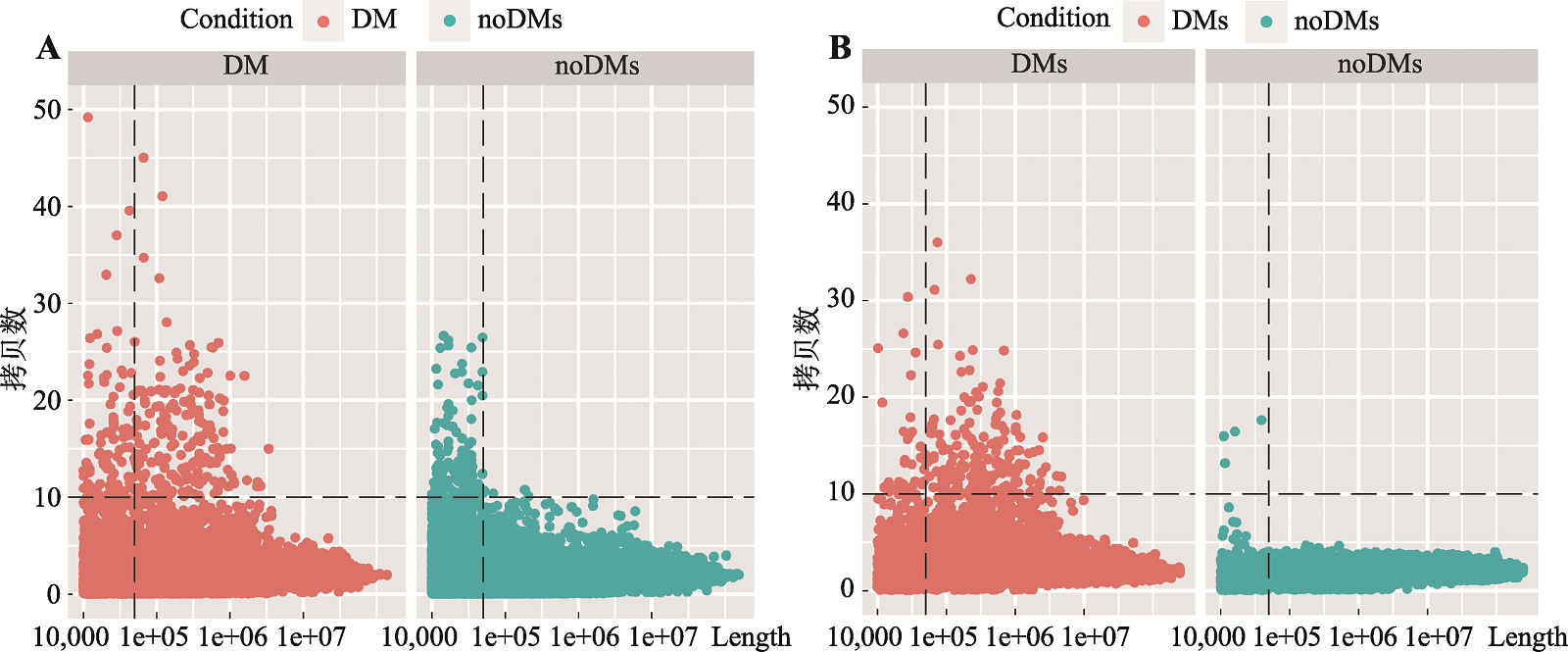
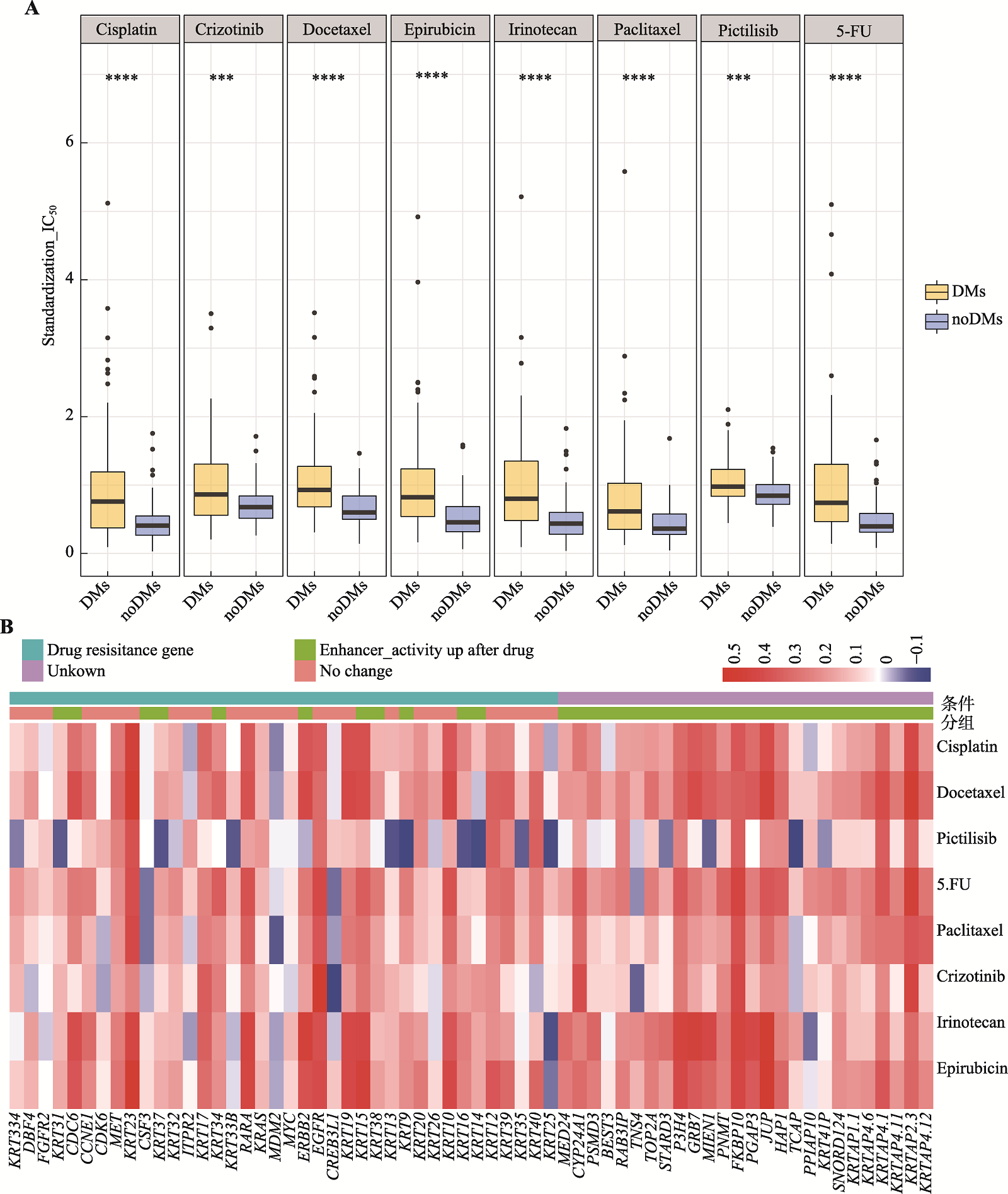
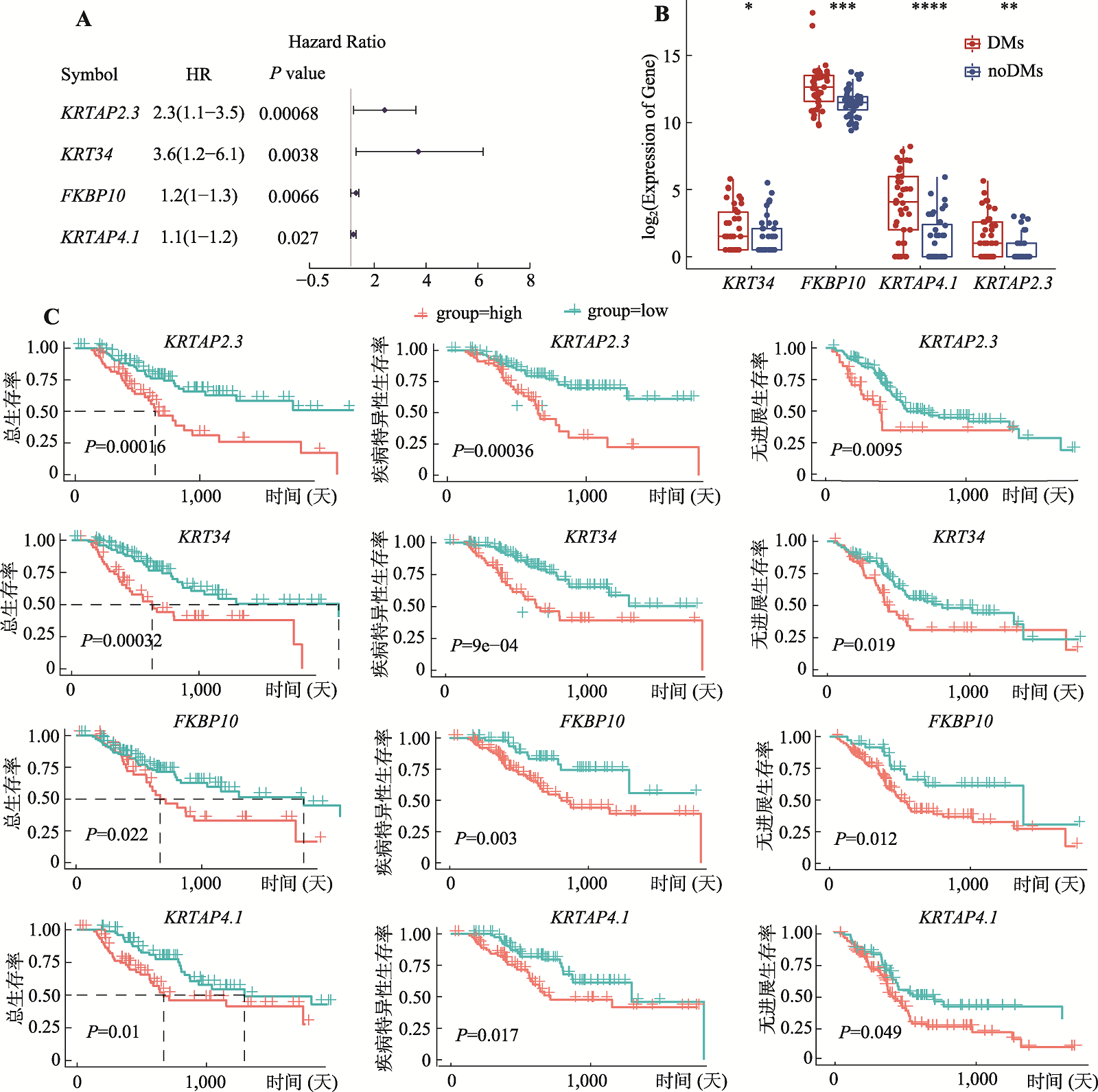
Analysis of regulatory mechanisms of enhancers in gastric cancer with double minute chromosomes based on bioinformatics
Mengting An1,2( ), Guanlin Guo1,2(
), Guanlin Guo1,2( ), Jie Wu1,2, Wenjing Sun1,2, Xueyuan Jia1,2(
), Jie Wu1,2, Wenjing Sun1,2, Xueyuan Jia1,2( )
)
- 1. Department of Medical Genetics, School of Bastic Medical Sciences, Harbin Medical University, Harbin 150081, China
2. Key Laboratory of Preservation of Human Genetic Resources and Disease Control in China (Harbin Medical University), Ministry of Education, Harbin 150081, China
-
Received:2024-12-17Revised:2025-02-04Online:2025-02-10Published:2025-02-10 -
Contact:Xueyuan Jia E-mail:anmenting814@163.com;ggl950913@163.com;jiaxueyuan@hrbmu.edu.cn -
Supported by:Natural Science Foundation of Heilongjiang Province of China(JQ2022C004);Joint Guiding Project of Heilongjiang Natural Science Foundation(LH2021H006);HMU Marshal Initiative Funding(HMUMIF-21007)
Cite this article
Mengting An, Guanlin Guo, Jie Wu, Wenjing Sun, Xueyuan Jia. Analysis of regulatory mechanisms of enhancers in gastric cancer with double minute chromosomes based on bioinformatics[J]. Hereditas(Beijing), 2025, 47(5): 558-572.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] |
Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2024, 74(3): 229-263.
pmid: 38572751 |
| [2] |
Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet, 2020, 396(10251): 635-648.
pmid: 32861308 |
| [3] |
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature, 2014, 513(7517): 202-209.
pmid: 25079317 |
| [4] |
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin, 2022, 72(1): 7-33.
pmid: 35020204 |
| [5] |
Sansregret L, Vanhaesebroeck B, Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat Rev Clin Oncol, 2018, 15(3): 139-150.
pmid: 29297505 |
| [6] |
Klein G, Klein E. Conditioned tumorigenicity of activated oncogenes. Cancer Res, 1986, 46(7): 3211-3224.
pmid: 3011242 |
| [7] |
Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet, 2003, 34(4): 369-376.
pmid: 12923544 |
| [8] |
Ilić M, Zaalberg IC, Raaijmakers JA, Medema RH. Life of double minutes: generation, maintenance, and elimination. Chromosoma, 2022, 131(3): 107-125.
pmid: 35487993 |
| [9] |
Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, Kornblum HI, Taylor MD, Kaushal S, Cavenee WK, Wechsler-Reya R, Furnari FB, Vandenberg SR, Rao PN, Wahl GM, Bafna V, Mischel PS. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature, 2017, 543(7643): 122-125.
pmid: 28178237 |
| [10] |
Kim H, Nguyen N-P, Turner K, Wu SH, Gujar AD, Luebeck J, Liu JH, Deshpande V, Rajkumar U, Namburi S, Amin SB, Yi E, Menghi F, Schulte JH, Henssen AG, Chang HY, Beck CR, Mischel PS, Bafna V, Verhaak RGW. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat Genet, 2020, 52(9): 891-897.
pmid: 32807987 |
| [11] |
Morton AR, Dogan-Artun N, Faber ZJ, MacLeod G, Bartels CF, Piazza MS, Allan KC, Mack SC, Wang XX, Gimple RC, Wu QL, Rubin BP, Shetty S, Angers S, Dirks PB, Sallari RC, Lupien M, Rich JN, Scacheri PC. Functional enhancers shape extrachromosomal oncogene amplifications. Cell, 2019, 179(6): 1330-1341.e13.
pmid: 31761532 |
| [12] |
Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet, 2011, 12(4): 283-293.
pmid: 21358745 |
| [13] | Liu Q, Li CY. The identification of enhancers and its application in cancer studies. Hereditas(Beijing), 2020, 42(9): 817-831. |
| 刘倩, 李春燕. 增强子的鉴定及其在肿瘤研究中的应用. 遗传, 2020, 42(9): 817-831. | |
| [14] |
Lu B, Zou CY, Yang ML, He YY, He JC, Zhang CX, Chen SY, Yu JM, Liu KY, Cao Q, Zhao W. Pharmacological inhibition of core regulatory circuitry liquid-liquid phase separation suppresses metastasis and chemoresistance in osteosarcoma. Adv Sci (Weinh), 2021, 8(20): e2101895.
pmid: 34432948 |
| [15] | Qi SH, Wang QL, Zhang JY, Liu Q, Li CY. The regulatory mechanisms by enhancers during cancer initiation and progression. Hereditas(Beijing), 2022, 44(4): 275-288. |
| 漆思晗, 王棨临, 张俊有, 刘倩, 李春燕. 增强子调控癌症发生发展的机制研究. 遗传, 2022, 44(4): 275-288. | |
| [16] |
Gupta PB, Fillmore CM, Jiang GZ, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell, 2011, 146(4): 633-644.
pmid: 21854987 |
| [17] |
Zhu YF, Gujar AD, Wong C-H, Tjong H, Ngan CY, Gong L, Chen YA, Kim H, Liu JH, Li MH, Mil-Homens A, Maurya R, Kuhlberg C, Sun FY, Yi E, deCarvalho AC, Ruan YJ, Verhaak RGW, Wei C-L. Oncogenic extrachromosomal DNA functions as mobile enhancers to globally amplify chromosomal transcription. Cancer Cell, 2021, 39(5): 694-707.e7.
pmid: 33836152 |
| [18] |
Lv HF, He YD, Nie CY, Du F, Chen XB. Adding of apatinib and camrelizumab to overcome de novo trastuzumab resistance of HER2-positive gastric cancer:a case report and literature review. Front Pharmacol, 2022, 13: 1067557.
pmid: 36699065 |
| [19] |
Huang PM, Ji FF, Cheung AHK, Fu KL, Zhou QM, Ding X, Chen DY, Lin YF, Wang LY, Jiao Y, Chu ESH, Kang W, To KF, Yu J, Wong CC. Peptostreptococcus stomatis promotes colonic tumorigenesis and receptor tyrosine kinase inhibitor resistance by activating ERBB2-MAPK. Cell Host Microbe, 2024, 32(8): 1365-1379.e10.
pmid: 39059397 |
| [20] |
Deshpande V, Luebeck J, Nguyen NPD, Bakhtiari M, Turner KM, Schwab R, Carter H, Mischel PS, Bafna V. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat Commun, 2019, 10(1): 392.
pmid: 30674876 |
| [21] |
Li QY, Tang HN, Hu FF, Qin CJ. Silencing of FOXO6 inhibits the proliferation, invasion, and glycolysis in colorectal cancer cells. J Cell Biochem, 2019, 120(3): 3853-3860.
pmid: 30321450 |
| [22] |
Song CL, Han Y, Luo H, Qin ZW, Chen ZQ, Liu Y, Lu S, Sun HM, Zhou CZ. HOXA10 induces BCL2 expression, inhibits apoptosis, and promotes cell proliferation in gastric cancer. Cancer Med, 2019, 8(12): 5651-5661.
pmid: 31364281 |
| [23] |
Li YM, Shi YK, Zhang XY, Li P, Ma L, Hu PB, Xu L, Dai YH, Xia S, Qiu H. FGFR2 upregulates PAI-1 via JAK2/STAT3 signaling to induce M2 polarization of macrophages in colorectal cancer. Biochim Biophys Acta Mol Basis Dis, 2023, 1869(4): 166665.
pmid: 36781088 |
| [24] |
Spitaleri G, Trillo Aliaga P, Attili I, Del Signore E, Corvaja C, Corti C, Uliano J, Passaro A, de Marinis F. MET in non-small-cell lung cancer (NSCLC): cross ‘a long and winding road’ looking for a target. Cancers (Basel), 2023, 15(19): 4779.
pmid: 37835473 |
| [25] |
Cai HQ, Zhang MJ, Cheng ZJ, Yu J, Yuan Q, Zhang J, Cai Y, Yang LY, Zhang Y, Hao JJ, Wang MR, Wan JH. FKBP10 promotes proliferation of glioma cells via activating AKT-CREB-PCNA axis. J Biomed Sci, 2021, 28(1): 13.
pmid: 33557829 |
| [26] |
Mori JO, White J, Elhussin I, Duduyemi BM, Karanam B, Yates C, Wang HH. Molecular and pathological subtypes related to prostate cancer disparities and disease outcomes in African American and European American patients. Front Oncol, 2022, 12: 928357.
pmid: 36033462 |
| [1] | Min Yang, Siyuan Lin, Changqi Yang, Yaosheng Chen, Zuyong He. Progress on SOX9 and its enhancers in mammalian sex determination [J]. Hereditas(Beijing), 2024, 46(9): 677-689. |
| [2] | Jilong Wang, Qing Li, Tingzheng Zhan. Principle and application of self-transcribing active regulatory region sequencing in enhancer discovery research [J]. Hereditas(Beijing), 2024, 46(8): 589-602. |
| [3] | Yao Chen, Xin Wen, Fangyuan Yuan, Chaoling Peng, Cuizhe Wang, Jun Zhang, Pingping Meng. Screening and validation of downstream target genes of SLC25A21 based on bioinformatics [J]. Hereditas(Beijing), 2024, 46(12): 1055-1065. |
| [4] | Xu Yan, Ying Guo, Donglin Sun, Nan Wu, Yan Jin. Drug resistance mechanism of anti-angiogenesis therapy in tumor [J]. Hereditas(Beijing), 2024, 46(11): 911-919. |
| [5] | Xiuli Chen, Haiyan Huang, Qiang Wu. Targeted deletion of 5′HS2 enhancer of β-globin locus control region in K562 cells [J]. Hereditas(Beijing), 2022, 44(9): 783-797. |
| [6] | Siyuan Xu, Jia Shou, Qiang Wu. Additional evidence of HS5-1 enhancer eRNA PEARL for protocadherin alpha gene regulation [J]. Hereditas(Beijing), 2022, 44(8): 695-764. |
| [7] | Sihan Qi, Qilin Wang, Junyou Zhang, Qian Liu, Chunyan Li. The regulatory mechanisms by enhancers during cancer initiation and progression [J]. Hereditas(Beijing), 2022, 44(4): 275-288. |
| [8] | Xingqi Wan, Wanzhen Wei, Shengliang Guo, Yixiao Cui, Xueying Jing, Lujie Huang, Jie Ma. Functional analysis of the long-range regulatory element of BMP2 gene [J]. Hereditas(Beijing), 2022, 44(12): 1141-1147. |
| [9] | Shanshan Wang, Wanyi Zhao, Huixiao Wu, Meng Shu, Jiaxin Yuan, Li Fang, Chao Xu. Research on the variants of FGFR1 and CEP290 genes in idiopathic hypogonadotropin hypogonadism [J]. Hereditas(Beijing), 2022, 44(10): 937-949. |
| [10] | Ling Wang, Jinhuan Li, Haiyan Huang, Qiang Wu. Serial deletions of tandem reverse CTCF sites reveal balanced HOXD regulatory landscape of enhancers [J]. Hereditas(Beijing), 2021, 43(8): 775-791. |
| [11] | Qian Liu, Chunyan Li. The identification of enhancers and its application in cancer studies [J]. Hereditas(Beijing), 2020, 42(9): 817-831. |
| [12] | Hong Xiang, Xiaohu Yang, Liangxia Ai, Yanping Pan, Yong Hu. Bioinformatics analysis of differentially expressed genes on alopecia [J]. Hereditas(Beijing), 2020, 42(2): 172-182. |
| [13] | Zhongyong Qin, Xiao Shi, Pingping Cao, Ying Chu, Wei Guan, Nan Yang, He Cheng, Yujie Sun. The NOXA promoter could function as an active enhancer to regulate the expression of BCL2 in the apoptosis response [J]. Hereditas(Beijing), 2020, 42(11): 1110-1121. |
| [14] | Hongqiang Lyu, Lele Hao, Erhu Liu, Zhifang Wu, Jiuqiang Han, Yuan Liu. Current status and future perspectives in bioinformatical analysis of Hi-C data [J]. Hereditas(Beijing), 2020, 42(1): 87-99. |
| [15] | Chao He,Wenlong Shen,Ping Li,Yan Zhang,Jing Zeng,Zuoming Yin,Zhihu Zhao. Bioinformatics analysis of Alu components at the level of genome 3D structure [J]. Hereditas(Beijing), 2019, 41(3): 254-261. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||