Hereditas(Beijing) ›› 2025, Vol. 47 ›› Issue (6): 660-671.doi: 10.16288/j.yczz.24-302
• Research Article • Previous Articles Next Articles
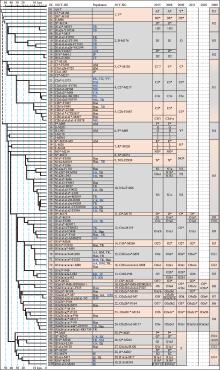
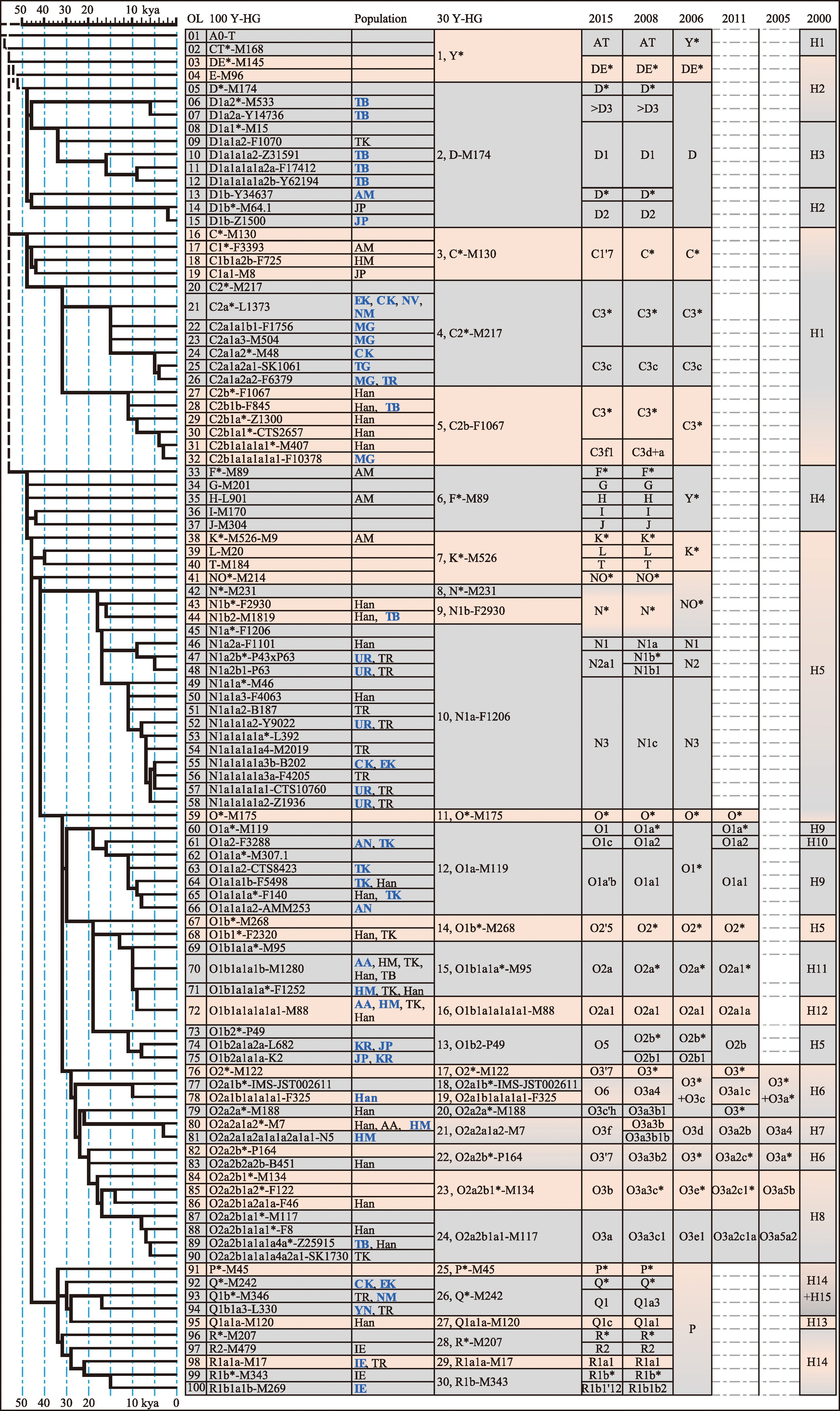
The 30−100 patrilineal nomenclature system for eastern Eurasian populations
Huixin Yu1( ), Xianpeng Zhang1(
), Xianpeng Zhang1( ), Lanhai Wei1,2(
), Lanhai Wei1,2( )
)
- 1. School of Ethnology and Anthropology, Inner Mongolia Normal University, Hohhot 021002, China
2. College of Life Science and Technology, Inner Mongolia Normal University, Hohhot 02100,China
-
Received:2024-12-14Revised:2025-04-13Online:2025-06-20Published:2025-04-14 -
Contact:Lanhai Wei E-mail:anthroaxiaoxin@163.com;jyzxp521@163.com;Ryan.lh.wei@foxmail.com -
Supported by:Project of Research Start-up Funds for High-level Talents Introduced by Inner Mongolia Normal University(2021RJRC002)
Cite this article
Huixin Yu, Xianpeng Zhang, Lanhai Wei. The 30−100 patrilineal nomenclature system for eastern Eurasian populations[J]. Hereditas(Beijing), 2025, 47(6): 660-671.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
| [1] | Wei LH, Li H, Jin L. Principles and applications of molecular anthropology. Shanghai: Shanghai Scientific and Technical Publishers, 2022, 1-16. |
| 韦兰海, 李辉, 金力. 分子人类学基本原理与应用. 上海: 上海科学技术出版社, 2022, 1-16. | |
| [2] | Ping WJ, Liu YC, Fu QM. Exploring the evolution of archaic humans through sedimentary ancient DNA. Hereditas (Beijing), 2022, 44(5): 362-369. |
| 平婉菁, 刘逸宸, 付巧妹. 沉积物古DNA探秘灭绝古人类演化. 遗传, 2022, 44(5):362-369. | |
| [3] | Yang QX, Wang MG, Liu C, Yuan HJ, He GL. Advancements and prospects in reconstructing the genetic genealogies of ancient and modern human populations using ancestral recombination graphs. Hereditas (Beijing), 2024, 46(10): 849-859. |
| 杨青鑫, 王萌鸽, 刘超, 袁慧军, 何光林. 基于祖先重组图重建古今人类群体遗传系谱的研究进展及展望. 遗传, 2024, 46(10): 849-859. | |
| [4] | Ping WJ, Xue JY, Fu QM. Ancient DNA elucidates the migration and evolutionary history of northern and southern populations in East Asia. Hereditas (Beijing), 2025, 47(1): 18-33. |
| 平婉菁, 薛家旸, 付巧妹. 古DNA解析东亚南北方人群的迁徙与演化历史. 遗传, 2025, 47(1): 18-33. | |
| [5] | Zhang DX, Dai SR, Cui YQ. The migration and evolutionary mechanisms of northern Asian populations from the perspective of ancient genomics. Hereditas (Beijing), 2025, 47(1): 34-45. |
| 张达轩, 戴沈汝, 崔银秋. 古基因组视角下的亚洲北部人群迁徙和演化机制. 遗传, 2025, 47(1): 34-45. | |
| [6] | Sun YH, Ping WJ, Liu YC, Fu QM. The ancient genome reveals the co-evolution history of human and lactic acid bacteria in the Bronze Age. Hereditas (Beijing), 2024, 46(11): 906-910. |
| 孙元宏, 平婉菁, 刘逸宸, 付巧妹. 古基因组揭示青铜时代人类与乳酸菌的协同演化史. 遗传, 2024, 46(11): 906-910. | |
| [7] | Wang HY, Hu YH, Cao YY, Zhu Q, Huang YG, Li X, Zhang J. AI-SNPs screening based on the whole genome data and research on genetic structure differences of subcontinent populations. Hereditas (Beijing), 2021, 43(10): 938-948. |
| 王浩宇, 胡渝涵, 曹悦岩, 朱强, 黄雨果, 李茜, 张霁. 基于全基因组数据的AI-SNPs筛选及大陆次级区域内群体遗传结构差异研究. 遗传, 2021, 43(10): 938-948. | |
| [8] | Wang XQ, Zhang QZ, Cheng P, Dong TT, Li WG, Zhou Z, Wang SQ. Genetic polymorphisms of 66 InDel loci in the Chinese Han population. Hereditas (Beijing), 2022, 44(4): 335-345. |
| 王雪倩, 张庆珍, 程鹏, 董婷婷, 李卫国, 周喆, 王升启. 中国汉族人群66个InDel基因座的遗传多态性. 遗传, 2022, 44(4): 335-345. | |
| [9] |
Fu QM, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PLF, Aximu-Petri A, Prüfer K, de Filippo C, Meyer M, Zwyns N, Salazar-García DC, Kuzmin YV, Keates SG, Kosintsev PA, Razhev DI, Richards MP, Peristov NV, Lachmann M, Douka K, Higham TFG, Slatkin M, Hublin JJ, Reich D, Kelso J, Viola TB, Pääbo S. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature, 2014, 514(7523): 445-449.
pmid: 25341783 |
| [10] |
Jónsson H, Sulem P, Kehr B, Kristmundsdottir S, Zink F, Hjartarson E, Hardarson MT, Hjorleifsson KE, Eggertsson HP, Gudjonsson SA, Ward LD, Arnadottir GA, Helgason EA, Helgason H, Gylfason A, Jonasdottir A, Jonasdottir A, Rafnar T, Frigge M, Stacey SN, Th Magnusson O, Thorsteinsdottir U, Masson G, Kong A, Halldorsson BV, Helgason A, Gudbjartsson DF, Stefansson K. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature, 2017, 549(7673): 519-522.
pmid: 28959963 |
| [11] |
Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res, 2008, 18(5): 830-838.
pmid: 18385274 |
| [12] |
Poznik GD, Xue YL, Mendez FL, Willems TF, Massaia A, Wilson Sayres MA, Ayub Q, McCarthy SA, Narechania A, Kashin S, Chen Y, Banerjee R, Rodriguez-Flores JL, Cerezo M, Shao HJ, Gymrek M, Malhotra A, Louzada S, Desalle R, Ritchie GRS, Cerveira E, Fitzgerald TW, Garrison E, Marcketta A, Mittelman D, Romanovitch M, Zhang CS, Zheng-Bradley XQ, Abecasis GR, McCarroll SA, Flicek P, Underhill PA, Coin L, Zerbino DR, Yang FT, Lee C, Clarke L, Auton A, Erlich Y, Handsaker RE, 1000 Genomes Project Consortium, Bustamante CD, Tyler-Smith C. Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat Genet, 2016, 48(6): 593-599.
pmid: 27111036 |
| [13] |
Liu MZ, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J, McGuire D, Tian C, Zhan XW, 23andMe Research Team, HUNT All-In Psychiatry, Choquet H, Docherty AR, Faul JD, Foerster JR, Fritsche LG, Gabrielsen ME, Gordon SD, Haessler J, Hottenga JJ, Huang HY, Jang SK, Jansen PR, Ling Y, Mägi R, Matoba N, McMahon G, Mulas A, Orrù V, Palviainen T, Pandit A, Reginsson GW, Skogholt AH, Smith JA, Taylor AE, Turman C, Willemsen G, Young H, Young KA, Zajac GJM, Zhao W, Zhou W, Bjornsdottir G, Boardman JD, Boehnke M, Boomsma DI, Chen C, Cucca F, Davies GE, Eaton CB, Ehringer MA, Esko T, Fiorillo E, Gillespie NA, Gudbjartsson DF, Haller T, Harris KM, Heath AC, Hewitt JK, Hickie IB, Hokanson JE, Hopfer CJ, Hunter DJ, Iacono WG, Johnson EO, Kamatani Y, Kardia SLR, Keller MC, Kellis M, Kooperberg C, Kraft P, Krauter KS, Laakso M, Lind PA, Loukola A, Lutz SM, Madden PAF, Martin NG, McGue M, McQueen MB, Medland SE, Metspalu A, Mohlke KL, Nielsen JB, Okada Y, Peters U, Polderman TJC, Posthuma D, Reiner AP, Rice JP, Rimm E, Rose RJ, Runarsdottir V, Stallings MC, Stančáková A, Stefansson H, Thai KK, Tindle HA, Tyrfingsson T, Wall TL, Weir DR, Weisner C, Whitfield JB, Winsvold BS, Yin J, Zuccolo L, Bierut LJ, Hveem K, Lee JJ, Munafò MR, Saccone NL, Willer CJ, Cornelis MC, David SP, Hinds DA, Jorgenson E, Kaprio J, Stitzel JA, Stefansson K, Thorgeirsson TE, Abecasis G, Liu DJJ, Vrieze S. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet, 2019, 51(2): 237-244.
pmid: 30643251 |
| [14] |
Underhill PA, Shen P, Lin AA, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ. Y chromosome sequence variation and the history of human populations. Nat Genet, 2000, 26(3): 358-361.
pmid: 11062480 |
| [15] |
Bajić V, Barbieri C, Hübner A, Güldemann T, Naumann C, Gerlach L, Berthold F, Nakagawa H, Mpoloka SW, Roewer L, Purps J, Stoneking M, Pakendorf B. Genetic structure and sex-biased gene flow in the history of southern African populations. Am J Phys Anthropol, 2018, 167(3): 656-671.
pmid: 30192370 |
| [16] |
Ke Y, Su B, Song X, Lu D, Chen L, Li H, Qi C, Marzuki S, Deka R, Underhill P, Xiao C, Shriver M, Lell J, Wallace D, Wells RS, Seielstad M, Oefner P, Zhu D, Jin J, Huang W, Chakraborty R, Chen Z, Jin L. African origin of modern humans in East Asia: a tale of 12,000 Y chromosomes. Science, 2001, 292(5519): 1151-1153.
pmid: 11349147 |
| [17] |
Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, Armenteros M, Arroyo E, Barbujani G, Beckman G, Beckman L, Bertranpetit J, Bosch E, Bradley DG, Brede G, Cooper G, Côrte-Real HB, de Knijff P, Decorte R, Dubrova YE, Evgrafov O, Gilissen A, Glisic S, Gölge M, Hill EW, Jeziorowska A, Kalaydjieva L, Kayser M, Kivisild T, Kravchenko SA, Krumina A, Kucinskas V, Lavinha J, Livshits LA, Malaspina P, Maria S, McElreavey K, Meitinger TA, Mikelsaar AV, Mitchell RJ, Nafa K, Nicholson J, Nørby S, Pandya A, Parik J, Patsalis PC, Pereira L, Peterlin B, Pielberg G, Prata MJ, Previderé C, Roewer L, Rootsi S, Rubinsztein DC, Saillard J, Santos FR, Stefanescu G, Sykes BC, Tolun A, Villems R, Tyler-Smith C, Jobling MA. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet, 2000, 67(6): 1526-1543.
pmid: 11078479 |
| [18] |
Kumar V, Reddy ANS, Babu JP, Rao TN, Langstieh BT, Thangaraj K, Reddy AG, Singh L, Reddy BM. Y-chromosome evidence suggests a common paternal heritage of Austro-Asiatic populations. BMC Evol Biol, 2007, 7: 47.
pmid: 17389048 |
| [19] | Wang F, Wang M, Zhang XH, Yu KL, Zheng LB, Yang YJ. Genetic substructure analysis of three isolated populations in southwest China. Hereditas (Beijing), 2022, 44(5): 424-431. |
| 王飞, 王萌, 张兴华, 宇克莉, 郑连斌, 杨亚军. 中国西南地区3个隔离人群遗传亚结构分析. 遗传, 2022, 44(5): 424-431. | |
| [20] | Zhu X, Jin X, Liu J, Yang L, Zou LX, Li CX, Huang J, Jiang L. Paternal genetic structure analysis of the modern Han populations based on Y-SNP and Y-STR. Hereditas (Beijing), 2024, 46(2): 149-167. |
| 朱信, 金鑫, 刘俊, 杨澜, 邹丽馨, 李彩霞, 黄江, 江丽. 基于Y-SNP和Y-STR揭示汉族人群父系遗传关系. 遗传, 2024, 46(2): 149-167. | |
| [21] | Zhang XP, Yu HX, Zhang J, Jia XY, He XF, Zhu BF, Wei LH, Yao HB. Multiple ancestral components and complex origins of the Yugur people in Gansu province revealing by 35 Y-STR. Hereditas (Beijing), 2024, 46(12): 1042-1054. |
| 张咸鹏, 于会新, 张劼, 贾欣怡, 何宵飞, 朱波峰, 韦兰海, 姚宏兵. 基于35 Y-STR研究甘肃裕固族的父系多重起源. 遗传, 2024, 46(12): 1042-1054. | |
| [22] |
Rasmussen M, Anzick SL, Waters MR, Skoglund P, DeGiorgio M, Stafford TW Jr, Rasmussen S, Moltke I, Albrechtsen A, Doyle SM, Poznik GD, Gudmundsdottir V, Yadav R, Malaspinas AS, White SS 5th, Allentoft ME, Cornejo OE, Tambets K, Eriksson A, Heintzman PD, Karmin M, Korneliussen TS, Meltzer DJ, Pierre TL, Stenderup J, Saag L, Warmuth VM, Lopes MC, Malhi RS, Brunak S, Sicheritz-Ponten T, Barnes I, Collins M, Orlando L, Balloux F, Manica A, Gupta R, Metspalu M, Bustamante CD, Jakobsson M, Nielsen R, Willerslev E. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature, 2014, 506(7487): 225-229.
pmid: 24522598 |
| [23] |
Helgason A, Einarsson AW, Guðmundsdóttir VB, Sigurðsson Á, Gunnarsdóttir ED, Jagadeesan A, Ebenesersdóttir SS, Kong A, Stefánsson K. The Y-chromosome point mutation rate in humans. Nat Genet, 2015, 47(5): 453-457.
pmid: 25807285 |
| [24] |
Y Chromosome Consortium. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res, 2002, 12(2): 339-348.
pmid: 11827954 |
| [25] |
Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet, 2003, 4(8): 598-612.
pmid: 12897772 |
| [26] |
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics, 2009, 25(16): 2078-2079.
pmid: 19505943 |
| [27] |
Shi H, Zhong H, Peng Y, Dong YL, Qi XB, Zhang F, Liu LF, Tan SJ, Ma RZ, Xiao CJ, Wells RS, Jin L, Su B. Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations. BMC Biol, 2008, 6: 45.
pmid: 18959782 |
| [28] |
Wen B, Xie XH, Gao S, Li H, Shi H, Song XF, Qian TZ, Xiao CJ, Jin JZ, Su B, Lu DR, Chakraborty R, Jin L. Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. Am J Hum Genet, 2004, 74(5): 856-865.
pmid: 15042512 |
| [29] | Sharengaowa, Ma PC, Yang WJ, Ochirbat A, Zhabagin M, Sun N, Xie YM, Li YL, Wei LH. Paternal origin of Mongolic-speaking populations: a review of studies from recent decades (1999-2019) and their implications for multidisciplinary research in the future. Hum Biol, 2023, 93(4): 269-288. |
| [30] |
Zhabagin MK, Sabitov Z, Tazhigulova I, Alborova I, Agdzhoyan A, Wei LH, Urasin V, Koshel S, Mustafin K, Akilzhanova A, Li H, Balanovsky O, Balanovska E. Medieval super-grandfather founder of western Kazakh clans from haplogroup C2a1a2-M48. J Hum Genet, 2021, 66(7): 707-716.
pmid: 33510364 |
| [31] |
Hudjashov G, Kivisild T, Underhill PA, Endicott P, Sanchez JJ, Lin AA, Shen P, Oefner P, Renfrew C, Villems R, Forster P. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc Natl Acad Sci USA, 2007, 104(21): 8726-8730.
pmid: 17496137 |
| [32] | Chen J, Li H, Qin ZD, Liu WH, Lin WX, Yin RX, Jin L, Pan SL. Y-chromosome genotyping and genetic structure of Zhuang populations. Acta Genetica Sinica, 2006, 33(12):1060-1072. |
| 陈晶, 李辉, 覃振东, 刘文泓, 林伟雄, 尹瑞兴, 金力, 潘尚领. 壮族Y染色体分型及其内部遗传结构. 遗传学报, 2006, 33(12): 1060-1072. | |
| [33] |
Zhang XM, Kampuansai J, Qi XB, Yan S, Yang ZH, Serey B, Sovannary T, Bunnath L, Aun HS, Samnom H, Kutanan W, Luo X, Liao SY, Kangwanpong D, Jin L, Shi H, Su B. An updated phylogeny of the human Y-chromosome lineage O2a-M95 with novel SNPs. PLoS One, 2014, 9(6): e101020.
pmid: 24972021 |
| [34] |
Rootsi S, Zhivotovsky LA, Baldovic M, Kayser M, Kutuev IA, Khusainova R, Bermisheva MA, Gubina M, Fedorova SA, Ilumäe AM, Khusnutdinova EK, Voevoda MI, Osipova LP, Stoneking M, Lin AA, Ferak V, Parik J, Kivisild T, Underhill PA, Villems R. A counter-clockwise northern route of the Y-chromosome haplogroup N from Southeast Asia towards Europe. Eur J Hum Genet, 2007, 15(2): 204-211.
pmid: 17149388 |
| [35] |
Karafet TM, Osipova LP, Gubina MA, Posukh OL, Zegura SL, Hammer MF. High levels of Y-chromosome differentiation among native Siberian populations and the genetic signature of a boreal hunter-gatherer way of life. Hum Biol, 2002, 74(6): 761-789.
pmid: 12617488 |
| [36] |
Wen B, Li H, Lu DR, Song XF, Zhang F, He YG, Li F, Gao Y, Mao XY, Zhang L, Qian J, Tan JZ, Jin JZ, Huang W, Deka R, Su B, Chakraborty R, Jin L. Genetic evidence supports demic diffusion of Han culture. Nature, 2004, 431(7006): 302-305.
pmid: 15372031 |
| [37] |
Deng W, Shi BC, He XL, Zhang ZH, Xu J, Li B, Yang J, Ling LJ, Dai CP, Qiang BQ, Shen Y, Chen RS. Evolution and migration history of the Chinese population inferred from Chinese Y-chromosome evidence. J Hum Genet, 2004, 49(7): 339-334.
pmid: 15173934 |
| [38] |
Derenko M, Malyarchuk B, Denisova GA, Wozniak M, Dambueva I, Dorzhu C, Luzina F, Miścicka-Sliwka D, Zakharov I. Contrasting patterns of Y-chromosome variation in South Siberian populations from Baikal and Altai-Sayan regions. Hum Genet, 2006, 118 (5): 591-604.
pmid: 16261343 |
| [39] |
Xue YL, Zerjal T, Bao WD, Zhu SL, Shu QF, Xu JJ, Du RF, Fu SB, Li P, Hurles ME, Yang HM, Tyler-Smith C. Male demography in East Asia: a north-south contrast in human population expansion times. Genetics, 2006, 172(4): 2431-2439.
pmid: 16489223 |
| [40] |
Kim SH, Kim KC, Shin DJ, Jin HJ, Kwak KD, Han MS, Song JM, Kim W, Kim W. High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea. Investig Genet, 2011, 2(1): 10.
pmid: 21463511 |
| [41] |
Hammer MF, Karafet TM, Park H, Omoto K, Harihara S, Stoneking M, Horai S. Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes. J Hum Genet, 2006, 51(1): 47-58.
pmid: 16328082 |
| [42] |
Lell JT, Sukernik RI, Starikovskaya YB, Su B, Jin L, Schurr TG, Underhill PA, Wallace DC. The dual origin and Siberian affinities of native American Y chromosomes. Am J Hum Genet, 2002, 70(1): 192-206.
pmid: 11731934 |
| [43] |
Malyarchuk BA, Derenko MV. Genetic history of the Koryaks and Evens of the Magadan region based on Y chromosome polymorphism data. Vavilovskii Zhurnal Genet Selektsii, 2024, 28(1): 90-97.
pmid: 38465253 |
| [44] |
Geppert M, Baeta M, Núñez C, Martínez-Jarreta B, Zweynert S, Cruz OWV, González-Andrade F, González- Solorzano J, Nagy M, Roewer L. Hierarchical Y-SNP assay to study the hidden diversity and phylogenetic relationship of native populations in South America. Forensic Sci Int Genet, 2011, 5(2): 100-104.
pmid: 20932815 |
| [45] |
Myres NM, Rootsi S, Lin AA, Järve M, King RJ, Kutuev I, Cabrera VM, Khusnutdinova EK, Pshenichnov A, Yunusbayev B, Balanovsky O, Balanovska E, Rudan P, Baldovic M, Herrera RJ, Chiaroni J, Di Cristofaro J, Villems R, Kivisild T, Underhill PA. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur J Hum Genet, 2011, 19(1): 95-101.
pmid: 20736979 |
| [46] |
Wang K, Prüfer K, Krause-Kyora B, Schuenemann VJ, Coia V, Maixner F, Zink A, Schiffels S, Krause J. High-coverage genome of the Tyrolean Iceman reveals unusually high Anatolian farmer ancestry. Cell Genom, 2023, 3(9): 100377.
pmid: 37719142 |
| [1] | Zuming Zhang, Hao Zhou, Xuezhi Huang, Duoyue Zhang, Jiayi Zhang, Yu Lin, Liwei Fang, Xiuchang Zhang, Yujun Cui, Yarong Wu, Yanjun Li. Study on population genomics of Bacillus anthracis based on multiple types of genetic variations [J]. Hereditas(Beijing), 2025, 47(6): 681-693. |
| [2] | Jiawen Ma, Xinle Liang. Analysis of structure and function of phage community occurring in the abnormal fermentation of vinegar mash through virome sequencing [J]. Hereditas(Beijing), 2025, 47(4): 489-498. |
| [3] | Can Liu, Weiwei Zhai, Xuemei Lu. Evolutionary ecology in tumor evolution: concept, application and innovation [J]. Hereditas(Beijing), 2025, 47(2): 228-236. |
| [4] | Xin Zhu, Xin Jin, Jun Liu, Lan Yang, Lixin Zou, Caixia Li, Jiang Huang, Li Jiang. Paternal genetic structure analysis of the modern Han populations based on Y-SNP and Y-STR [J]. Hereditas(Beijing), 2024, 46(2): 149-167. |
| [5] | Fei Wang, Meng Wang, Xinghua Zhang, Keli Yu, Lianbin Zheng, Yajun Yang. Genetic substructure analysis of three isolated populations in southwest China [J]. Hereditas(Beijing), 2022, 44(5): 424-431. |
| [6] | Wanjing Ping, Yichen Liu, Qiaomei Fu. Exploring the evolution of archaic humans through sedimentary ancient DNA [J]. Hereditas(Beijing), 2022, 44(5): 362-369. |
| [7] | Xueqian Wang, Qingzhen Zhang, Peng Cheng, Tingting Dong, Weiguo Li, Zhe Zhou, Shengqi Wang. Genetic polymorphisms of 66 InDel loci in the Chinese Han population [J]. Hereditas(Beijing), 2022, 44(4): 335-345. |
| [8] | Hongyuan Zheng, Lin Yan, Chao Yang, Yarong Wu, Jingliang Qin, Tongyu Hao, Dajin Yang, Yunchang Guo, Xiaoyan Pei, Tongyan Zhao, Yujun Cui. Population genomics study of Vibrio alginolyticus [J]. Hereditas(Beijing), 2021, 43(4): 350-361. |
| [9] | Zilong Wen, Yiqiang Zhao. Progress on animal domestication under population genetics [J]. Hereditas(Beijing), 2021, 43(3): 226-239. |
| [10] | Haoyu Wang, Yuhan Hu, Yueyan Cao, Qiang Zhu, Yuguo Huang, Xi Li, Ji Zhang. AI-SNPs screening based on the whole genome data and research on genetic structure differences of subcontinent populations [J]. Hereditas(Beijing), 2021, 43(10): 938-948. |
| [11] | Zhiyong Liu, He Ren, Chong Chen, Jingjing Zhang, Xiaomeng Zhang, Yan Shi, Linyu Shi, Ying Chen, Feng Cheng, Li Jia, Man Chen, Qingwei Fan, Jiarong Zhang, Wanting Li, Mengchun Wang, Zilin Ren, Yacheng Liu, Ming Ni, Hongyu Sun, Jiangwei Yan. Actual mutational research of 19 autosomal STRs based on restricted mutation model and big data [J]. Hereditas(Beijing), 2021, 43(10): 949-961. |
| [12] | Xingdong Chen, Yanfeng Jiang, Ping Xu, Li Jin. Construction and utilization of human genetic resources in large population cohorts [J]. Hereditas(Beijing), 2021, 43(10): 980-987. |
| [13] | Wenxiu Wang, Tao Huang, Liming Li. Construction and application of human genetic resources in the China Kadoorie Biobank [J]. Hereditas(Beijing), 2021, 43(10): 972-979. |
| [14] | Ming Liu, Yi Li, Yafang Yang, Yuwen Yan, Fan Liu, Caixia Li, Faming Zeng, Wenting Zhao. Human facial shape related SNP analysis in Han Chinese populations [J]. Hereditas(Beijing), 2020, 42(7): 680-690. |
| [15] | Xiaojuan Wang, Enfang Qian, Yue Li, Zhengyang Song, Hui Zhao, Hexin Xie, Caixia Li, Jiang Huang, Li Jiang. A genetic sub-structure study of the Tibetan population in Southwest China [J]. Hereditas(Beijing), 2020, 42(6): 565-576. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||