Hereditas(Beijing) ›› 2024, Vol. 46 ›› Issue (2): 149-167.doi: 10.16288/j.yczz.23-260
• Research Article • Previous Articles Next Articles
Paternal genetic structure analysis of the modern Han populations based on Y-SNP and Y-STR
Xin Zhu1,2( ), Xin Jin3, Jun Liu2,4, Lan Yang2,4, Lixin Zou2,5, Caixia Li1,2, Jiang Huang1(
), Xin Jin3, Jun Liu2,4, Lan Yang2,4, Lixin Zou2,5, Caixia Li1,2, Jiang Huang1( ), Li Jiang2(
), Li Jiang2( )
)
- 1. Institute of Forensic Medicine, Guizhou Medical University, Guiyang 550004, China
2. Key Laboratory of Forensic Genetics, Beijing Engineering Research Center of Crime Scene Evidence Examination, National Engineering Laboratory for Forensic Science, Institute of Forensic Science, Beijing 100038, China
3. Public Security Department of Hainan Province, Haikou 570203, China
4. Shanxi Medical University, Taiyuan 030001, China
5. Jiangsu Normal University, Xuzhou 221116, China
-
Received:2023-12-11Revised:2024-01-24Online:2024-02-20Published:2024-01-26 -
Contact:Jiang Huang,Li Jiang E-mail:1987538437@qq.com;mmm_hj@gmc.edu.cn;jl@mail.bnu.edu.cn -
Supported by:National Key R&D Program of China(2022YFC3341004);National Natural Science Foundation of China(82171870);Fundamental Research Funds for Institute of Forensic Science(2022JB020);Open Project of the National Engineering Laboratory for Forensic Science(2021FGKFKT01)
Cite this article
Xin Zhu, Xin Jin, Jun Liu, Lan Yang, Lixin Zou, Caixia Li, Jiang Huang, Li Jiang. Paternal genetic structure analysis of the modern Han populations based on Y-SNP and Y-STR[J]. Hereditas(Beijing), 2024, 46(2): 149-167.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
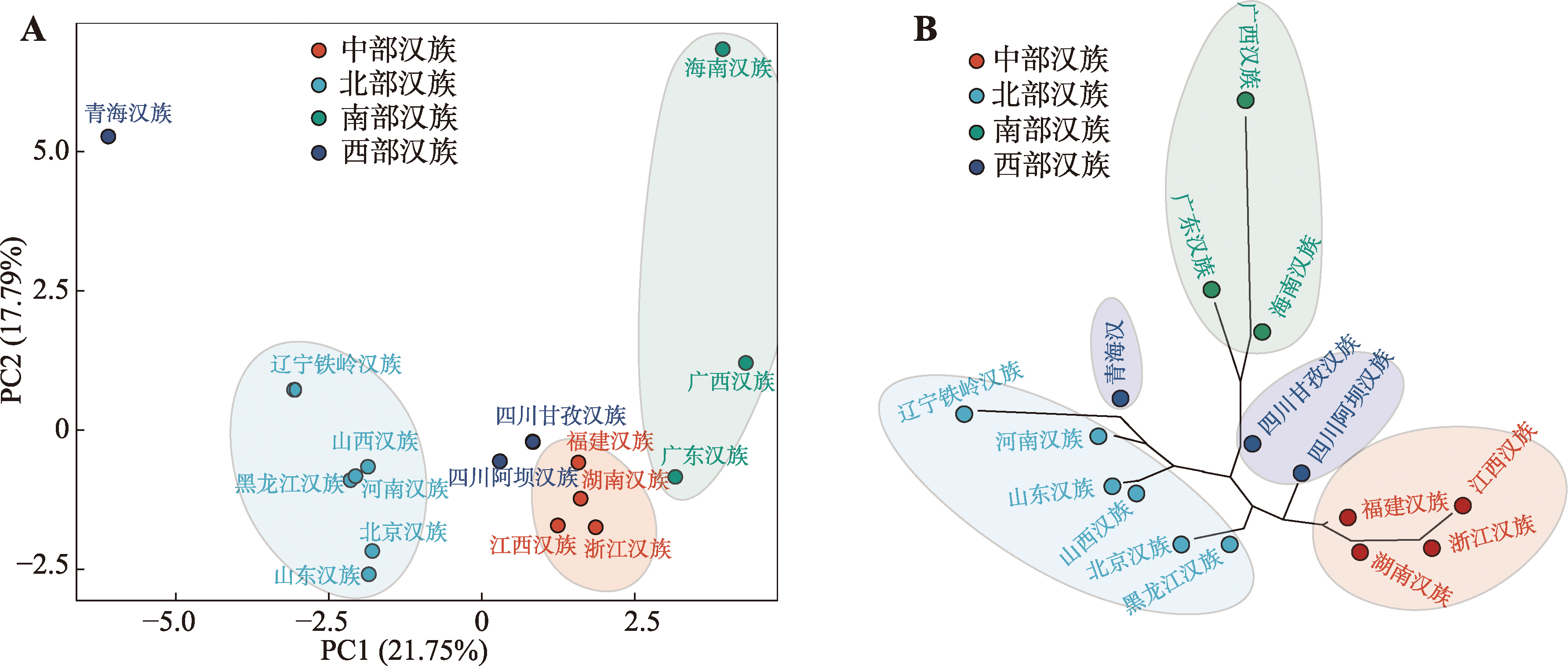
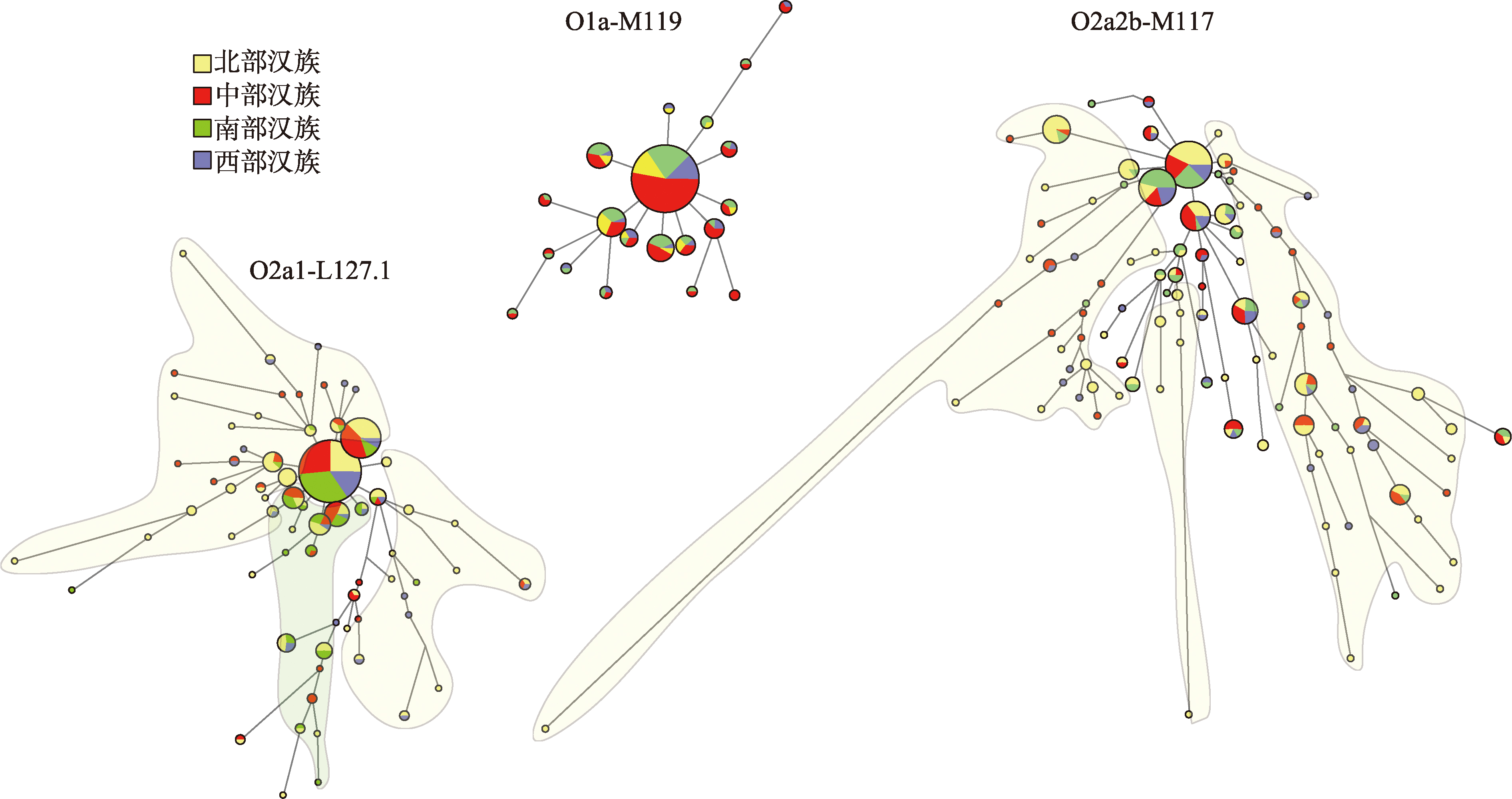
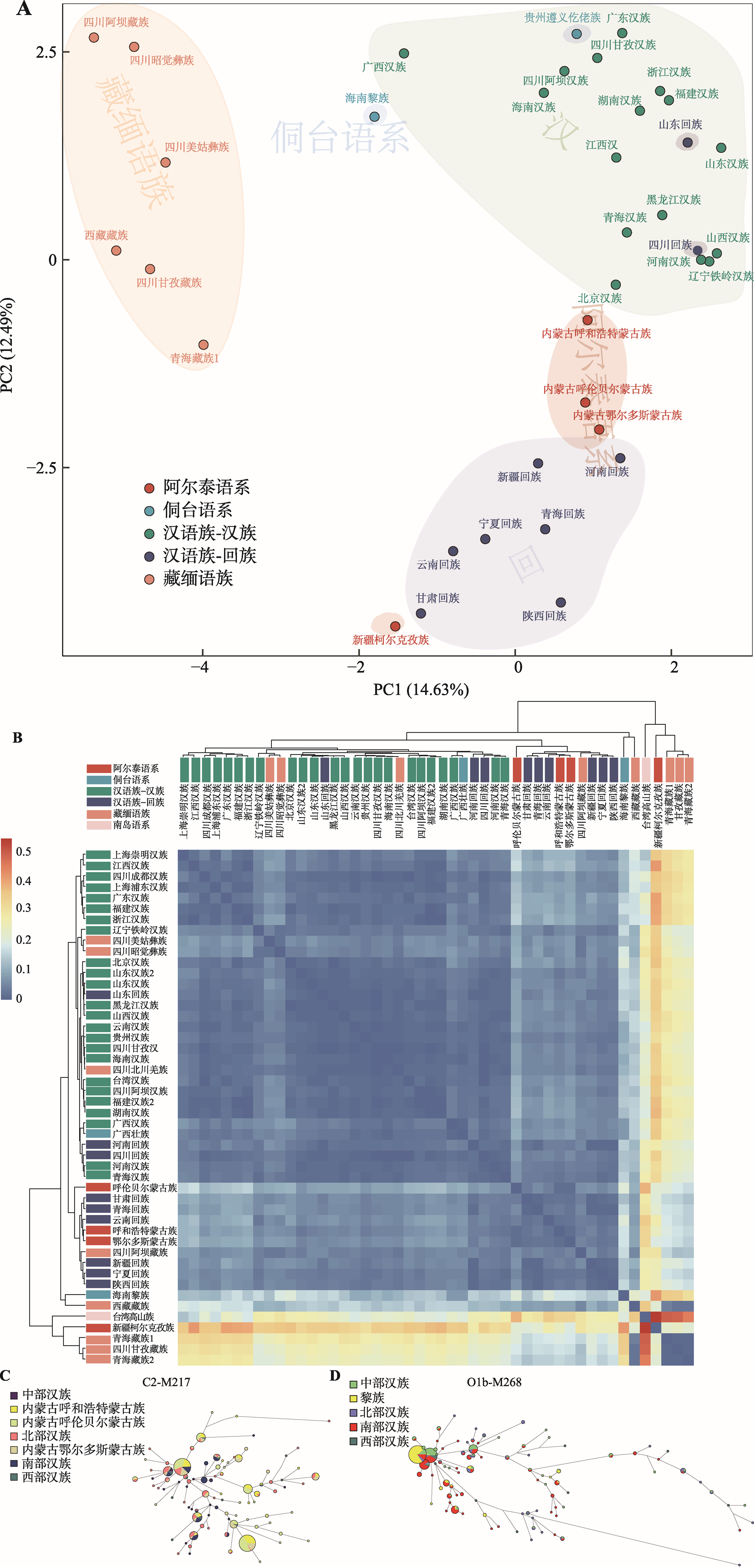
Table 1
List of chinese population information"
| 序号 | 语系 | 语族 | 群体 | 样本量 | 数据集 | 来源 |
|---|---|---|---|---|---|---|
| 1 | 汉藏语系 | 汉语族 | 辽宁铁岭汉族 | 80 | 研究人群 | 自检 |
| 2 | 黑龙江汉族 | 94 | 研究人群 | 文献[ | ||
| 3 | 北京汉族 | 114 | 研究人群 | 文献[ | ||
| 4 | 山西汉族 | 99 | 研究人群 | 文献[ | ||
| 5 | 山东汉族 | 96 | 研究人群 | 文献[ | ||
| 6 | 河南汉族 | 199 | 研究人群 | 文献[ | ||
| 7 | 江西汉族 | 110 | 研究人群 | 文献[ | ||
| 8 | 湖南汉族 | 106 | 研究人群 | 文献[ | ||
| 9 | 福建汉族 | 124 | 研究人群 | 文献[ | ||
| 10 | 浙江汉族 | 97 | 研究人群 | 文献[ | ||
| 11 | 青海汉族 | 114 | 研究人群 | 自检 | ||
| 12 | 四川阿坝汉族 | 76 | 研究人群 | 自检 | ||
| 13 | 四川甘孜汉族 | 91 | 研究人群 | 自检 | ||
| 14 | 广东汉族 | 117 | 研究人群 | 文献[ | ||
| 15 | 广西汉族 | 113 | 研究人群 | 文献[ | ||
| 16 | 海南汉族 | 200 | 研究人群 | 文献[ | ||
| 17 | 新疆回族 | 49 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 18 | 青海回族 | 69 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 19 | 甘肃回族 | 167 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 20 | 宁夏回族 | 85 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 21 | 陕西回族 | 84 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 22 | 河南回族 | 67 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 23 | 山东回族 | 48 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 24 | 四川回族 | 38 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 25 | 云南回族 | 43 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 26 | 藏缅语族 | 四川甘孜藏族 | 88 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | |
| 27 | 青海藏族1 | 82 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 28 | 西藏藏族 | 251 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 29 | 四川阿坝藏族 | 68 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 30 | 四川美姑彝族 | 222 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 31 | 四川昭觉彝族 | 205 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 32 | 四川北川羌族 | 422 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 33 | 阿尔泰语系 | 蒙古语族 | 内蒙古呼和浩特蒙古族 | 228 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ |
| 34 | 内蒙古呼伦贝尔蒙古族 | 217 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 35 | 内蒙古鄂尔多斯蒙古族 | 199 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | ||
| 36 | 突厥语族 | 新疆柯尔克孜族 | 297 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ | |
| 37 | 侗台语系 | 侗水语族 | 海南黎族 | 102 | 60Y-SNP数据集,16Y-STR数据集 | 文献[ |
| 38 | 仡央语族 | 贵州遵义仡佬族 | 68 | 60Y-SNP数据集 | 文献[ | |
| 39 | 汉藏语系 | 汉语族 | 四川成都汉族 | 2228 | 16Y-STR数据集 | 文献[ |
| 40 | 福建汉族2 | 50 | 16Y-STR数据集 | 文献[ | ||
| 41 | 台湾汉族 | 93 | 16Y-STR数据集 | 文献[ | ||
| 42 | 上海崇明岛汉族 | 527 | 16Y-STR数据集 | 文献[ | ||
| 43 | 上海浦东汉族 | 689 | 16Y-STR数据集 | 文献[ | ||
| 44 | 山东汉族2 | 303 | 16Y-STR数据集 | 文献[ | ||
| 45 | 云南汉族 | 540 | 16Y-STR数据集 | 文献[ | ||
| 46 | 藏缅语族 | 青海藏族2 | 163 | 16Y-STR数据集 | 文献[ | |
| 47 | 侗台语系 | 壮傣语族 | 广西壮族 | 201 | 16Y-STR数据集 | 文献[ |
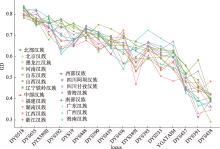
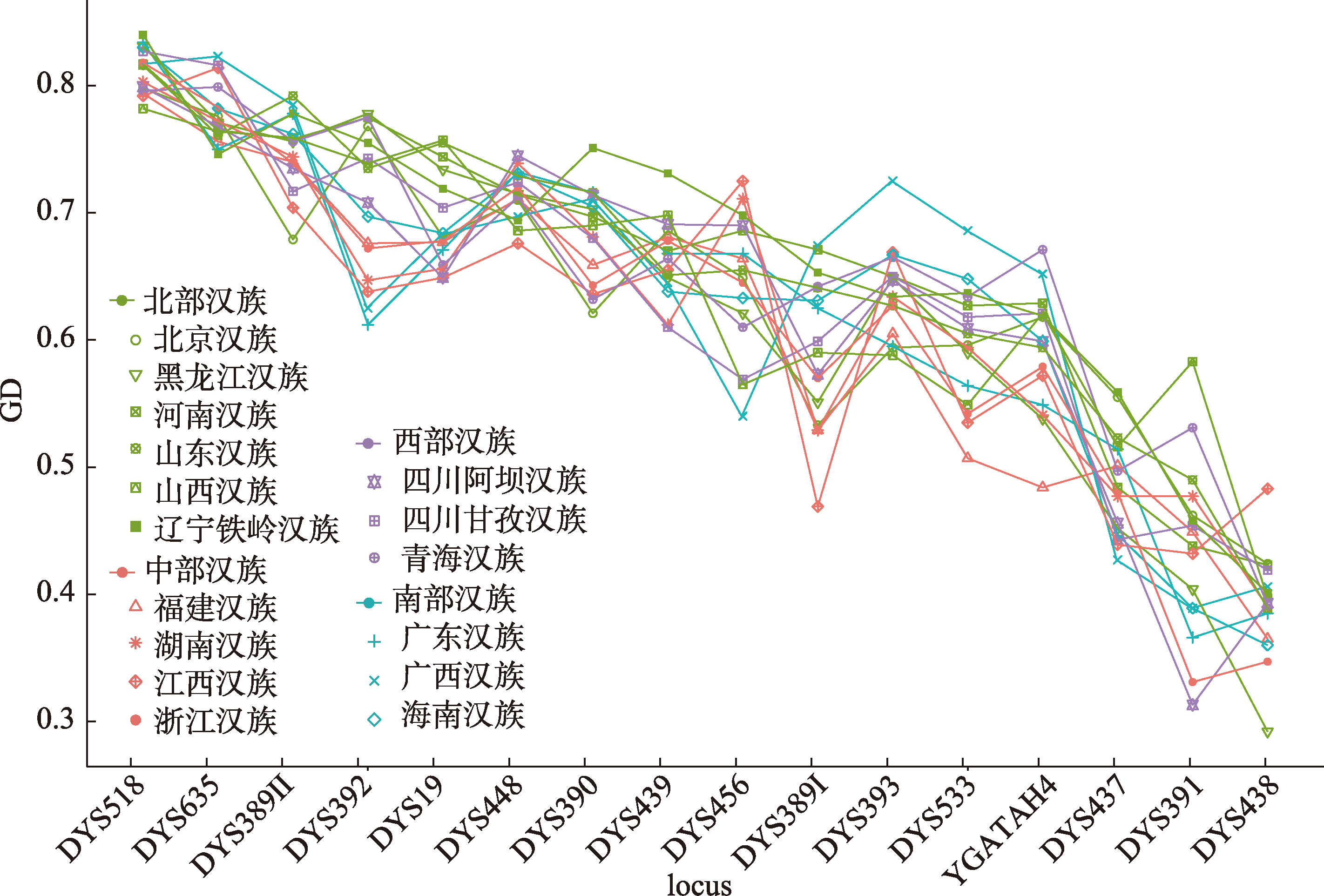
Table 2
Forensic parameters of Han populations based on 16 Y-STR loci"
| 地区 | 样本量 | 人群 | 观察到的单倍型数量 | |||
|---|---|---|---|---|---|---|
| 单倍型数 | MP | HD | DC | |||
| 中国北部 | 114 | 北京汉族 | 113 | 0.00908 | 0.99968 | 0.99123 |
| 94 | 黑龙江汉族 | 94 | 0.01064 | 1 | 1 | |
| 80 | 辽宁铁岭汉族 | 80 | 0.01250 | 1 | 1 | |
| 199 | 河南汉族 | 195 | 0.00544 | 0.99958 | 0.97990 | |
| 96 | 山东汉族 | 95 | 0.01086 | 0.99955 | 0.98958 | |
| 99 | 山西汉族 | 99 | 0.01010 | 1 | 1 | |
| 中国西部 | 114 | 青海汉族 | 113 | 0.00932 | 0.99937 | 0.98261 |
| 76 | 四川阿坝汉族 | 75 | 0.01387 | 0.99928 | 0.98684 | |
| 91 | 四川甘孜汉族 | 88 | 0.01214 | 0.99896 | 0.97778 | |
| 中国中部 | 106 | 湖南汉族 | 104 | 0.01017 | 0.99926 | 0.98113 |
| 110 | 江西汉族 | 108 | 0.00977 | 0.99931 | 0.98182 | |
| 97 | 浙江汉族 | 95 | 0.01119 | 0.99911 | 0.97938 | |
| 124 | 福建汉族 | 116 | 0.01040 | 0.99764 | 0.93548 | |
| 中国南部 | 117 | 广东汉族 | 115 | 0.00915 | 0.99939 | 0.98291 |
| 113 | 广西汉族 | 110 | 0.00983 | 0.99901 | 0.97345 | |
| 200 | 海南汉族 | 195 | 0.00552 | 0.99947 | 0.97500 | |
| [1] |
Jobling MA, Tyler-Smith C. Human Y-chromosome variation in the genome-sequencing era. Nat Rev Genet, 2017, 18(8): 485-497.
doi: 10.1038/nrg.2017.36 pmid: 28555659 |
| [2] |
Batini C, Jobling MA. Detecting past male-mediated expansions using the Y chromosome. Hum Genet, 2017, 136(5): 547-557.
doi: 10.1007/s00439-017-1781-z pmid: 28349239 |
| [3] |
King TE, Jobling MA. Founders, drift, and infidelity: the relationship between Y chromosome diversity and patrilineal surnames. Mol Biol Evol, 2009, 26(5): 1093-1102.
doi: 10.1093/molbev/msp022 pmid: 19204044 |
| [4] |
Charchar FJ, Bloomer LD, Barnes TA, Cowley MJ, Nelson CP, Wang Y, Denniff M, Debiec R, Christofidou P, Nankervis S, Dominiczak AF, Bani-Mustafa A, Balmforth AJ, Hall AS, Erdmann J, Cambien F, Deloukas P, Hengstenberg C, Packard C, Schunkert H, Ouwehand WH, Ford I, Goodall AH, Jobling MA, Samani NJ, Tomaszewski M. Inheritance of coronary artery disease in men: an analysis of the role of the Y chromosome. Lancet, 2012, 379(9819): 915-922.
doi: S0140-6736(11)61453-0 pmid: 22325189 |
| [5] | Khan K, Siddiqi MH, Abbas M, Almas M, Idrees M. Forensic applications of Y chromosomal properties. Leg Med (Tokyo), 2017, (26): 86-91. |
| [6] |
Kayser M. Forensic use of Y-chromosome DNA: a general overview. Hum Genet, 2017, 136(5): 621-635.
doi: 10.1007/s00439-017-1776-9 pmid: 28315050 |
| [7] | Santos FR, Epplen JT, Pena SD. Testing deficiency paternity cases with a Y-linked tetranucleotide repeat polymorphism. Exs, 1993, (67): 261-265. |
| [8] |
Corach D, Filgueira Risso L, Marino M, Penacino G, Sala A. Routine Y-STR typing in forensic casework. Forensic Sci Int, 2001, 118(2-3): 131-135.
pmid: 11311825 |
| [9] | Pinto N, Gusmão L, Amorim A. Mutation and mutation rates at Y chromosome specific Short Tandem Repeat Polymorphisms (STRs): a reappraisal. Forensic Sci Int Genet, 2014, (9): 20-24. |
| [10] |
Laouina A, Nadifi S, Boulouiz R, El Arji M, Talbi J, El Houate B, Yahia H, Chbel F. Mutation rate at 17 Y-STR loci in "Father/Son" pairs from moroccan population. Leg Med (Tokyo), 2013, 15(5): 269-271.
doi: 10.1016/j.legalmed.2013.03.003 pmid: 23623014 |
| [11] |
Ballantyne KN, Goedbloed M, Fang RX, Schaap O, Lao O, Wollstein A, Choi Y, van Duijn K, Vermeulen M, Brauer S, Decorte R, Poetsch M, von Wurmb-Schwark N, de Knijff P, Labuda D, Vézina H, Knoblauch H, Lessig R, Roewer L, Ploski R, Dobosz T, Henke L, Henke J, Furtado MR, Kayser M. Mutability of Y-chromosomal microsatellites: rates, characteristics, molecular bases, and forensic implications. Am J Hum Genet, 2010, 87(3): 341-353.
doi: 10.1016/j.ajhg.2010.08.006 pmid: 20817138 |
| [12] |
Claerhout S, Vandenbosch M, Nivelle K, Gruyters L, Peeters A, Larmuseau MHD, Decorte R. Determining Y-STR mutation rates in deep-routing genealogies: Identification of haplogroup differences. Forensic Sci Int Genet, 2018, 34: 1-10.
doi: 10.1016/j.fsigen.2018.01.005 |
| [13] | Wickenheiser RA. Expanding DNA database effectiveness. Forensic Sci Int Synergy, 2022, 4: 100226. |
| [14] |
Xue YL, Wang QJ, Long Q, Ng BL, Swerdlow H, Burton J, Skuce C, Taylor R, Abdellah Z, Zhao YL, Asan, MacArthur DG, Quail MA, Carter NP, Yang HM, Tyler-Smith C. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr Biol, 2009, 19(17): 1453- 1457.
doi: 10.1016/j.cub.2009.07.032 pmid: 19716302 |
| [15] |
Wang CC, Li H. Inferring human history in East Asia from Y chromosomes. Investig Genet, 2013, 4(1): 11.
doi: 10.1186/2041-2223-4-11 |
| [16] |
Yoshida Y, Kubo S. Y-SNP and Y-STR analysis in a Japanese population. Leg Med (Tokyo), 2008, 10(5): 243-252.
doi: 10.1016/j.legalmed.2008.01.008 pmid: 18378482 |
| [17] |
Cortellini V, Verzeletti A, Cerri N, Marino A, De Ferrari F,. Y-chromosome polymorphisms and ethnic group—a combined STR and SNP approach in a population sample from northern Italy. Croat Med J, 2013, 54(3): 279-285.
pmid: 23771759 |
| [18] | Song MY, Song F, Zhao CX, Hou YP. YHP: Y-chromosome Haplogroup Predictor for predicting male lineages based on Y-STRs. bioRxiv, 2021. |
| [19] |
Yin CY, Sun H, Zhou HG, Jin L, Li SL. EA-YPredictor: one new software developed to predict pedigree haplogroup based on Y-STR haplotypes. Forensic Science and Technology, 2020, 45(2): 117-124.
doi: 10.16467/j.1008-3650.2020.02.002 |
|
殷才湧, 孙辉, 周怀谷, 金立, 李士林. EA-YPredictor:基于Y-STR数据的家系特异性单倍群归属判别分析软件. 刑事技术, 2020, 45(2): 117-124.
doi: 10.16467/j.1008-3650.2020.02.002 |
|
| [20] |
Dogan S, Babic N, Gurkan C, Goksu A, Marjanovic D, Hadziavdic V. Y-chromosomal haplogroup distribution in the Tuzla Canton of Bosnia and Herzegovina: a concordance study using four different in silico assignment algorithms based on Y-STR data. Homo, 2016, 67(6): 471-483.
doi: S0018-442X(16)30059-2 pmid: 27908490 |
| [21] |
Ralf A, Van Oven M, Zhong KY, Kayser M. Simultaneous analysis of hundreds of Y-chromosomal SNPs for high-resolution paternal lineage classification using targeted semiconductor sequencing. Hum Mutat, 2015, 36(1): 151-159.
doi: 10.1002/humu.22713 pmid: 25338970 |
| [22] |
Xie MK, Song F, Li JN, Lang M, Luo HB, Wang Z, Wu J, Li CZ, Tian CC, Wang WZ, Ma H, Song Z, Fan YJ, Hou YP. Genetic substructure and forensic characteristics of Chinese Hui populations using 157 Y-SNPs and 27 Y-STRs. Forensic Sci Int Genet, 2019, 41: 11-18.
doi: 10.1016/j.fsigen.2019.03.022 |
| [23] |
Wang MG, Wang Z, He GL, Liu J, Wang SY, Qian XQ, Lang M, Li JN, Xie MK, Li CT, Hou YP. Developmental validation of a custom panel including 165 Y-SNPs for Chinese Y-chromosomal haplogroups dissection using the ion S5 XL system. Forensic Sci Int Genet, 2019, 38: 70-76.
doi: 10.1016/j.fsigen.2018.10.009 |
| [24] |
Yin CY, Ren YJ, Adnan A, Tian JZ, Guo KJ, Xia MY, He ZW, Zhai D, Chen XY, Wang L, Li X, Qin XJ, Li SL, Jin L. Title: developmental validation of Y-SNP pedigree tagging system: a panel via quick ARMS PCR. Forensic Sci Int Genet, 2020, 46: 102271.
doi: 10.1016/j.fsigen.2020.102271 |
| [25] |
Zhou ZH, Zhou YX, Yao YN, Qian JL, Liu BN, Yang QR, Shao CC, Li H, Sun K, Tang QQ, Xie JH. A 16-plex Y-SNP typing system based on allele-specific PCR for the genotyping of Chinese Y-chromosomal haplogroups. Leg Med (Tokyo), 2020, 46: 101720.
doi: S1344-6223(20)30054-7 pmid: 32505804 |
| [26] |
Claerhout S, Verstraete P, Warnez L, Vanpaemel S, Larmuseau M, Decorte R. CSYseq: the first Y-chromosome sequencing tool typing a large number of Y-SNPs and Y-STRs to unravel worldwide human population genetics. PLoS Genet, 2021, 17(9): e1009758.
doi: 10.1371/journal.pgen.1009758 |
| [27] |
Tao RY, Li M, Chai SY, Xia RC, Qu YL, Yuan CL, Yang GY, Dong XY, Bian YN, Zhang SH, Li CT. Developmental validation of a 381 Y-chromosome SNP panel for haplogroup analysis in the Chinese populations. Forensic Sci Int Genet, 2023, 62: 102803.
doi: 10.1016/j.fsigen.2022.102803 |
| [28] |
He GL, Wang MG, Miao L, Chen J, Zhao J, Sun QX, Duan SH, Wang ZY, Xu XF, Sun YT, Liu Y, Liu J, Wang Z, Wei LH, Liu C, Ye J, Wang L. Multiple founding paternal lineages inferred from the newly-developed 639-plex Y-SNP panel suggested the complex admixture and migration history of Chinese people. Hum Genomics, 2023, 17(1): 29.
doi: 10.1186/s40246-023-00476-6 pmid: 36973821 |
| [29] |
Liu J, Jiang LR, Zhao MY, Du WA, Wen YF, Li SY, Zhang SY, Fang FF, Shen J, He GL, Wang MG, Dai H, Hou YP, Wang Z. Development and validation of a custom panel including 256 Y-SNPs for Chinese Y-chromosomal haplogroups dissection. Forensic Sci Int Genet, 2022, 61: 102786.
doi: 10.1016/j.fsigen.2022.102786 |
| [30] | 中华人民共和国国家统计局.国家数据-2021年度第七次人口普查. (2021-11-26)[2023-10-11].http://www.stats.gov.cn/sj/pcsj/rkpc/d7c/. |
| [31] |
Yu XE, Li H. Origin of ethnic groups, linguistic families, and civilizations in China viewed from the Y chromosome. Mol Genet Genomics, 2021, 296(4): 783-797.
doi: 10.1007/s00438-021-01794-x |
| [32] |
Li YC, Ye WJ, Jiang CG, Zeng Z, Tian JY, Yang LQ, Liu KJ, Kong QP. River valleys shaped the maternal genetic landscape of Han Chinese. Mol Biol Evol, 2019, 36(8): 1643-1652.
doi: 10.1093/molbev/msz072 |
| [33] |
Lang M, Liu H, Song F, Qiao XH, Ye Y, Ren H, Li JN, Huang J, Xie MK, Chen SJ, Song MY, Zhang YF, Qian XQ, Yuan TX, Wang Z, Liu YM, Wang MG, Liu YC, Liu J, Hou YP. Forensic characteristics and genetic analysis of both 27 Y-STRs and 143 Y-SNPs in Eastern Han Chinese population. Forensic Sci Int Genet, 2019, 42: e13-e20.
doi: 10.1016/j.fsigen.2019.07.011 |
| [34] | Gu JQ, Jiang L, Xu JY, Wang H, Wei YL, Li CX. Genetic structure of East Asians based on high-density SNP data. Prog Biochem Biophys, 2023, 50(11): 1-14. |
| 顾佳琪, 江丽, 徐景怡, 王寒, 魏以梁, 李彩霞. 基于高密度SNP数据的东亚人群遗传结构研究. 生物化学与生物物理进展, 2023, 50(11): 1-14. | |
| [35] | Zhang XY, Lu L, Yu H, Zhang X, Deng HB. Research on the distribution characteristics and formation mechanisms of Chinese traditional villages. World Regional Studies, 2023, 32(4): 132-143. |
| 张晓瑶, 陆林, 虞虎, 张潇, 邓洪波. 中国传统村落分布特征与成因机制研究. 世界地理研究, 2023, 32(4): 132-143. | |
| [36] |
Chiang CWK, Mangul S, Robles C, Sankararaman S. A comprehensive map of genetic variation in the world's largest ethnic group-Han Chinese. Mol Biol Evol, 2018, 35(11): 2736-2750.
doi: 10.1093/molbev/msy170 pmid: 30169787 |
| [37] |
Xu SH, Yin XY, Li SL, Jin WF, Lou HY, Yang L, Gong XH, Wang HY, Shen YP, Pan XD, He YG, Yang YJ, Wang Y, Fu WQ, An Y, Wang JC, Tan JZ, Qian J, Chen XL, Zhang X, Sun YF, Zhang XJ, Wu BL, Jin L. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet, 2009, 85(6): 762-774.
doi: 10.1016/j.ajhg.2009.10.015 pmid: 19944404 |
| [38] |
Cao YN, Li L, Xu M, Feng ZM, Sun XH, Lu JL, Xu Y, Du PN, Wang TG, Hu RY, Ye Z, Shi LX, Tang XL, Yan L, Gao ZN, Chen G, Zhang YF, Chen LL, Ning G, Bi YF, Wang WQ, ChinaMAP Consortium. The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals. Cell Res, 2020, 30(9): 717-731.
doi: 10.1038/s41422-020-0322-9 pmid: 32355288 |
| [39] |
Yin CY, Su KY, He ZW, Zhai D, Guo KJ, Chen XY, Jin L, Li SL. Genetic reconstruction and forensic analysis of Chinese Shandong and Yunnan Han Populations by co-analyzing Y Chromosomal STRs and SNPs. Genes (Basel), 2020, 11(7): 743.
doi: 10.3390/genes11070743 |
| [40] |
Zhang X, Tang Z, Wang B, Zhou XD, Zhou LM, Zhang GY, Tian JZ, Zhao YQ, Yao ZQ, Tian L, Zhang SH, Xia H, Jin L, Li CT, Li SL. Forensic analysis and genetic structure construction of Chinese Chongming Island Han based on Y Chromosome STRs and SNPs. Genes (Basel), 2022, 13(8): 1363.
doi: 10.3390/genes13081363 |
| [41] |
Zhang YL, Zhang R, Li M, Luo L, Zhang JY, Ding JD, Zhang SH, Li CT, Bian YN, Zhou CJ. Genetic polymorphism of both 29 Y-STRs and 213 Y-SNPs in Han populations from Shandong Province, China. Leg Med (Tokyo), 2020, 47:101738.
doi: S1344-6223(20)30072-9 pmid: 32818903 |
| [42] |
Song MY, Wang Z, Zhang YQ, Zhao CX, Lang M, Xie MK, Qian XQ, Wang MG, Hou YP. Forensic characteristics and phylogenetic analysis of both Y-STR and Y-SNP in the Li and Han ethnic groups from Hainan Island of China. Forensic Sci Int Genet, 2019, 39: e14-e20.
doi: 10.1016/j.fsigen.2018.11.016 |
| [43] |
Song F, Song MY, Luo HB, Xie MK, Wang XD, Dai H, Hou YP.Paternal genetic structure of Kyrgyz ethnic group in China revealed by high-resolution Y-chromosome STRs and SNPs. Electrophoresis, 2021, 42(19): 1892-1899.
doi: 10.1002/elps.202100142 pmid: 34169540 |
| [44] |
Wang MG, He GL, Zou X, Liu J, Ye ZW, Ming TY, Du WA, Wang Z, Hou YP. Genetic insights into the paternal admixture history of Chinese Mongolians via high- resolution customized Y-SNP SNaPshot panels. Forensic Sci Int Genet, 2021, 54: 102565.
doi: 10.1016/j.fsigen.2021.102565 |
| [45] |
Song MY, Wang ZF, Lyu Q, Ying J, Wu Q, Jiang LR, Wang F, Zhou YX, Song F, Luo HB, Hou YP, Song XB, Ying BW. Paternal genetic structure of the Qiang ethnic group in China revealed by high-resolution Y-chromosome STRs and SNPs. Forensic Sci Int Genet, 2022, 61: 102774.
doi: 10.1016/j.fsigen.2022.102774 |
| [46] |
Wang F, Song F, Song MY, Li JN, Xie MK, Hou YP. Genetic reconstruction and phylogenetic analysis by 193 Y-SNPs and 27 Y-STRs in a Chinese Yi ethnic group. Electrophoresis, 2021, 42(14-15): 1480-1487.
doi: 10.1002/elps.202100003 pmid: 33909307 |
| [47] |
Wang F, Song F, Song MY, Luo HB, Hou YP. Genetic structure and paternal admixture of the modern Chinese Zhuang population based on 37 Y-STRs and 233 Y-SNPs. Forensic Sci Int Genet, 2022, 58: 102681.
doi: 10.1016/j.fsigen.2022.102681 |
| [48] | Wang XJ, Qian EF, Li Y, Song ZY, Zhao H, Xie HX, Li CX, Huang J, Jiang L. A genetic sub-structure study of the Tibetan population in Southwest China. Hereditas (Beijing), 2020, 42(6): 565-576. |
| 王小娟, 钱恩芳, 李悦, 宋正阳, 赵慧, 谢何鑫, 李彩霞, 黄江, 江丽. 中国西南地区藏族人群遗传亚结构研究. 遗传, 2020, 42(6): 565-576. | |
| [49] |
Wang XJ, Jiang L, Qian EF, Long F, Cui W, Ji AQ, Zhang FS, Zou K, Huang J, Li CX. Genetic polymorphisms and haplotypic structure analysis of the Guizhou Gelao ethnic group based on 35 Y-STR loci. Leg Med (Tokyo), 2020, 43: 101666.
doi: S1344-6223(19)30333-5 pmid: 31972469 |
| [50] |
Trejaut JA, Poloni ES, Yen JC, Lai YH, Loo JH, Lee CL, He CL, Lin M. Taiwan Y-chromosomal DNA variation and its relationship with Island Southeast Asia. BMC Genet, 2014, 15: 77.
doi: 10.1186/1471-2156-15-77 pmid: 24965575 |
| [51] |
Wang H, Mao J, Xia Y, Bai XG, Zhu WQ, Peng D, Liang WB. Genetic polymorphisms of 17 Y-chromosomal STRs in the Chengdu Han population of China. Int J Legal Med, 2017, 131(4): 967-968.
doi: 10.1007/s00414-016-1511-4 pmid: 27942858 |
| [52] |
Zhu BF, Wu YM, Shen CM, Yang TH, Deng YJ, Xun X, Tian YF, Yan JC, Li T. Genetic analysis of 17 Y-chromosomal STRs haplotypes of Chinese Tibetan ethnic group residing in Qinghai province of China. Forensic Sci Int, 2008, 175(2-3): 238-243.
doi: 10.1016/j.forsciint.2007.06.012 pmid: 17659855 |
| [53] | ISOGG. Y-DNA haplogroup tree. (2019-10-08)[2022-11-22].https://isogg.org/tree/index.html. |
| [54] |
Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA, 1973, 70(12): 3321-3323.
doi: 10.1073/pnas.70.12.3321 pmid: 4519626 |
| [55] |
Nei M, Tajima F. DNA polymorphism detectable by restriction endonucleases. Genetics, 1981, 97(1): 145-163.
doi: 10.1093/genetics/97.1.145 pmid: 6266912 |
| [56] | Ito K, Murphy D. Application of ggplot2 to pharmacometric graphics. CPT Pharmacometrics Syst Pharmacol, 2013, 2(10): e79. |
| [57] |
Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour, 2010, 10(3): 564-567.
doi: 10.1111/j.1755-0998.2010.02847.x pmid: 21565059 |
| [58] |
Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics, 1992, 131(2): 479-491.
doi: 10.1093/genetics/131.2.479 pmid: 1644282 |
| [59] |
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol, 2018, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [60] |
Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol, 1999, 16(1): 37-48.
doi: 10.1093/oxfordjournals.molbev.a026036 pmid: 10331250 |
| [61] | 蔡晓云.Y染色体揭示的早期人类进入东亚和东亚人群特征形成过程[学位论文]. 复旦大学, 2009. |
| [62] |
Yan S, Wang CC, Zheng HX, Wang W, Qin ZD, Wei LH, Wang Y, Pan XD, Fu WQ, He YG, Xiong LJ, Jin WF, Li SL, An Y, Li H, Jin L. Y chromosomes of 40% Chinese descend from three Neolithic super-grandfathers. PLoS One, 2014, 9(8): e105691.
doi: 10.1371/journal.pone.0105691 |
| [63] |
Sagart L, Jacques G, Lai YF, Ryder RJ, Thouzeau V, Greenhill SJ, List JM. Dated language phylogenies shed light on the ancestry of Sino-Tibetan. Proc Natl Acad Sci USA, 2019, 116(21): 10317-10322.
doi: 10.1073/pnas.1817972116 pmid: 31061123 |
| [64] |
Wang LX, Lu Y, Zhang C, Wei LH, Yan S, Huang YZ, Wang CC, Mallick S, Wen SQ, Jin L, Xu SH, Li H. Reconstruction of Y-chromosome phylogeny reveals two neolithic expansions of Tibeto-Burman populations. Mol Genet Genomics, 2018, 293(5): 1293-1300.
doi: 10.1007/s00438-018-1461-2 |
| [65] |
Wang CC, Yeh HY, Popov AN, Zhang HQ, Matsumura H, Sirak K, Cheronet O, Kovalev A, Rohland N, Kim AM, Mallick S, Bernardos R, Tumen D, Zhao J, Liu YC, Liu JY, Mah M, Wang K, Zhang Z, Adamski N, Broomandkhoshbacht N, Callan K, Candilio F, Carlson KSD, Culleton BJ, Eccles L, Freilich S, Keating D, Lawson AM, Mandl K, Michel M, Oppenheimer J, Özdoğan KT, Stewardson K, Wen SQ, Yan S, Zalzala F, Chuang R, Huang CJ, Looh H, Shiung CC, Nikitin YG, Tabarev AV, Tishkin AA, Lin S, Sun ZY, Wu XM, Yang TL, Hu X, Chen L, Du H, Bayarsaikhan J, Mijiddorj E, Erdenebaatar D, Iderkhangai TO, Myagmar E, Kanzawa-Kiriyama H, Nishino M, Shinoda KI, Shubina OA, Guo JX, Cai WW, Deng QY, Kang LL, Li DW, Li DN, Lin R, Nini, Shrestha R, Wang LX, Wei LH, Xie GM, Yao HB, Zhang MF, He GL, Yang XM, Hu R, Robbeets M, Schiffels S, Kennett DJ, Jin L, Li H, Krause J, Pinhasi R, Reich D. Genomic insights into the formation of human populations in East Asia. Nature, 2021, 591(7850): 413-419.
doi: 10.1038/s41586-021-03336-2 |
| [66] |
Zhong H, Shi H, Qi XB, Duan ZY, Tan PP, Jin L, Su B, Ma RZ. Extended Y chromosome investigation suggests postglacial migrations of modern humans into East Asia via the northern route. Mol Biol Evol, 2011, 28(1): 717-727.
doi: 10.1093/molbev/msq247 pmid: 20837606 |
| [67] |
Shi H, Dong YL, Wen B, Xiao CJ, Underhill PA, Shen PD, Chakraborty R, Jin L, Su B.Y-chromosome evidence of southern origin of the East Asian-specific haplogroup O3-M122. Am J Hum Genet, 2005, 77(3): 408-419.
doi: 10.1086/444436 pmid: 16080116 |
| [68] |
Ning C, Yan S, Hu K, Cui YQ, Jin L.Refined phylogenetic structure of an abundant East Asian Y-chromosomal haplogroup O*-M134. Eur J Hum Genet, 2016, 24(2): 307-309.
doi: 10.1038/ejhg.2015.183 pmid: 26306641 |
| [69] |
Kang LL, Lu Y, Wang CC, Hu K, Chen F, Liu K, Li SL, Jin L, Li H, Genographic Consortium. Y-chromosome O 3 haplogroup diversity in Sino-Tibetan populations reveals two migration routes into the eastern Himalayas. Ann Hum Genet, 2012, 76(1): 92-99.
doi: 10.1111/ahg.2012.76.issue-1 |
| [70] |
Zhang XM, Kampuansai J, Qi XB, Yan S, Yang ZH, Serey B, Sovannary T, Bunnath L, Aun HS, Samnom H, Kutanan W, Luo X, Liao SY, Kangwanpong D, Jin L, Shi H, Su B. An updated phylogeny of the human Y-chromosome lineage O2a-M95 with novel SNPs. PLoS One, 2014, 9(6): e101020.
doi: 10.1371/journal.pone.0101020 |
| [71] |
Sun J, Li YX, Ma PC, Yan S, Cheng HZ, Fan ZQ, Deng XH, Ru K, Wang CC, Wang CC, Chen G, Wei LH. Shared paternal ancestry of Han, Tai-Kadai-speaking and Austronesian-speaking populations as revealed by the high resolution phylogeny of O1a-M119 and distribution of its sub-lineages within China. Am J Phys Anthropol, 2021, 174(4): 686-700.
doi: 10.1002/ajpa.v174.4 |
| [72] |
Yan S, Wang CC, Li H, Li SL, Jin L, Genographic Consortium. An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4. Eur J Hum Genet, 2011, 19(9): 1013-1015.
doi: 10.1038/ejhg.2011.64 pmid: 21505448 |
| [73] |
Chen H, Lin R, Lu Y, Zhang R, Gao Y, He YG, Xu SH. Tracing Bai-Yue ancestry in aboriginal Li People on Hainan Island. Mol Biol Evol, 2022, 39(10): msac210.
doi: 10.1093/molbev/msac210 |
| [74] |
Wei LH, Huang YZ, Yan S, Wen SQ, Wang LX, Du PX, Yao DL, Li SL, Yang YJ, Jin L, Li H. Phylogeny of Y-chromosome haplogroup C3b-F1756, an important paternal lineage in Altaic-speaking populations. J Hum Genet, 2017, 62(10): 915-918.
doi: 10.1038/jhg.2017.60 |
| [75] |
Zhabagin M, Sabitov Z, Tazhigulova I, Alborova I, Agdzhoyan A, Wei LH, Urasin V, Koshel S, Mustafin K, Akilzhanova A, Li H, Balanovsky O, Balanovska E.Medieval super-grandfather founder of Western Kazakh Clans from Haplogroup C2a1a2-M48. J Hum Genet, 2021, 66(7): 707-716.
doi: 10.1038/s10038-021-00901-5 pmid: 33510364 |
| [76] |
Dulik MC, Osipova LP, Schurr TG. Y-chromosome variation in Altaian Kazakhs reveals a common paternal gene pool for Kazakhs and the influence of Mongolian expansions. PLoS One, 2011, 6(3): e17548.
doi: 10.1371/journal.pone.0017548 |
| [77] |
Chiaroni J, Underhill PA, Cavalli-Sforza LL. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc Natl Acad Sci USA, 2009, 106(48): 20174-20179.
doi: 10.1073/pnas.0910803106 pmid: 19920170 |
| [78] |
Qian EF, Deng P, Huang MS, Ma Q, Zhao H, Li CX, Huang J, Jiang L. Genetic polymorphism of Y chromosome haplogroup D-M174 in East Asian populations. J Forensic Med, 2019, 35(3): 308-313.
doi: 10.12116/j.issn.1004-5619.2019.03.009 |
|
钱恩芳, 邓盼, 黄美莎, 马泉, 赵慧, 李彩霞, 黄江, 江丽. 东亚族群中Y染色体D-M174单倍群的遗传多态性. 法医学杂志, 2019, 35(3): 308-313.
doi: 10.12116/j.issn.1004-5619.2019.03.009 |
|
| [79] |
Shi H, Zhong H, Peng Y, Dong YL, Qi XB, Zhang F, Liu LF, Tan SJ, Ma RZ, Xiao CJ, Wells RS, Jin L, Su B. Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations. BMC biology, 2008, 6: 45.
doi: 10.1186/1741-7007-6-45 pmid: 18959782 |
| [80] |
Balaresque P, Bowden GR, Parkin EJ, Omran GA, Heyer E, Quintana-Murci L, Roewer L, Stoneking M, Nasidze I, Carvalho-Silva DR, Tyler-Smith C, de Knijff P, Jobling MA. Dynamic nature of the proximal AZFc region of the human Y chromosome: multiple independent deletion and duplication events revealed by microsatellite analysis. Hum Mutat, 2008, 29(10): 1171-1180.
doi: 10.1002/humu.20757 pmid: 18470947 |
| [81] |
Malyarchuk B, Derenko M, Denisova G, Wozniak M, Grzybowski T, Dambueva I, Zakharov I. Phylogeography of the Y-chromosome haplogroup C in northern Eurasia. Ann Hum Genet, 2010, 74(6): 539-546.
doi: 10.1111/j.1469-1809.2010.00601.x pmid: 20726964 |
| [82] |
Tofanelli S, Ferri G, Bulayeva K, Caciagli L, Onofri V, Taglioli L, Bulayev O, Boschi I, Alù M, Berti A, Rapone C, Beduschi G, Luiselli D, Cadenas AM, Awadelkarim KD, Mariani-Costantini R, Elwali NE, Verginelli F, Pilli E, Herrera RJ, Gusmão L, Paoli G, Capelli C. J1-M267 Y lineage marks climate-driven pre-historical human displacements. Eur J Hum Genet, 2009, 17(11): 1520-1524.
doi: 10.1038/ejhg.2009.58 pmid: 19367321 |
| [83] |
Myres NM, Ekins JE, Lin AA, Cavalli-Sforza LL, Woodward SR, Underhill PA.Y-chromosome short tandem repeat DYS458.2 non-consensus alleles occur independently in both binary haplogroups J1-M267 and R1b3-M405. Croat Med J, 2007, 48(4): 450-459.
pmid: 17696299 |
| [84] |
Wang CC, Lu Y, Kang LL, Ding HQ, Yan S, Guo JX, Zhang Q, Wen SQ, Wang LX, Zhang MF, Tong XZ, Huang XF, Nie SJ, Deng QY, Zhu BF, Jin L, Li H. The massive assimilation of indigenous East Asian populations in the origin of Muslim Hui people inferred from paternal Y chromosome. Am J Phys Anthropol, 2019, 169(2): 341-347.
doi: 10.1002/ajpa.v169.2 |
| [85] |
Wang QY, Zhao J, Ren Z, Sun J, He GL, Guo JX, Zhang HL, Ji JY, Liu YB, Yang MQ, Yang XM, Chen JW, Zhu KY, Wang R, Li YX, Chen G, Huang J, Wang CC. Male-dominated migration and massive mssimilation of indigenous East Asians in the formation of muslim Hui People in Southwest China. Front Genet, 2020, 11: 618614.
doi: 10.3389/fgene.2020.618614 |
| [86] |
Zhong H, Shi H, Qi XB, Xiao CJ, Jin L, Ma RZ, Su B. Global distribution of Y-chromosome haplogroup C reveals the prehistoric migration routes of African exodus and early settlement in East Asia. J Hum Genet, 2010, 55(7): 428-435.
doi: 10.1038/jhg.2010.40 pmid: 20448651 |
| [1] | Xin Li, Hong Fan, Xingchun Zhao, Xiaonuo Fan, Ruoxia Yao. Rapid analyzing mixed STR profiles based on the global minimum residual method [J]. Hereditas(Beijing), 2023, 45(10): 933-944. |
| [2] | Fei Wang, Meng Wang, Xinghua Zhang, Keli Yu, Lianbin Zheng, Yajun Yang. Genetic substructure analysis of three isolated populations in southwest China [J]. Hereditas(Beijing), 2022, 44(5): 424-431. |
| [3] | Yongqiang Kong, Jinkai Liu, Jiaqi Gu, Jingyi Xu, Yunuo Zheng, Yiliang Wei, Shaoyuan Wu. Optimization scheme of machine learning model for genetic division between northern Han, southern Han, Korean and Japanese [J]. Hereditas(Beijing), 2022, 44(11): 1028-1043. |
| [4] | Xiaoyuan Guo, Changchun Sun, Siyao Xue, Hui Zhao, Li Jiang, Caixia Li. 49AISNP: a study on the ancestry inference of the three ethnic groups in the north of East Asia [J]. Hereditas(Beijing), 2021, 43(9): 880-889. |
| [5] | Zilong Wen, Yiqiang Zhao. Progress on animal domestication under population genetics [J]. Hereditas(Beijing), 2021, 43(3): 226-239. |
| [6] | Xi Li, Haoyu Wang, Yueyan Cao, Qiang Zhu, Panyin Shu, Tingyun Hou, Yuting Wang, Ji Zhang. Forensic genomics research on microhaplotypes [J]. Hereditas(Beijing), 2021, 43(10): 962-971. |
| [7] | Zhiyong Liu, Riga Wu, Ran Li, Qiangwei Wang, Hongyu Sun. Ethical issues of the research and practice in forensic genetics [J]. Hereditas(Beijing), 2021, 43(10): 994-1002. |
| [8] | Xiaojuan Wang, Enfang Qian, Yue Li, Zhengyang Song, Hui Zhao, Hexin Xie, Caixia Li, Jiang Huang, Li Jiang. A genetic sub-structure study of the Tibetan population in Southwest China [J]. Hereditas(Beijing), 2020, 42(6): 565-576. |
| [9] | Yang Liu, Changchun Sun, Mi Ma, Ling Wang, Wenting Zhao, Quan Ma, Anquan Ji, Jing Liu, Caixia Li. The ancestry inference of Chinese populations using 74-plex SNPs system [J]. Hereditas(Beijing), 2020, 42(3): 296-308. |
| [10] | Jian Hu,Yiren Zhou,Jialin Ding,Zhiyuan Wang,Ling Liu,Yekai Wang,Huiling Lou,Shouyi Qiao,Yanhua Wu. Simplification of genotyping techniques of the ABO blood type experiment and exploration of population genetics [J]. Hereditas(Beijing), 2017, 39(5): 423-429. |
| [11] | Li Jiang,Qifan Sun,Quan Ma,Wenting Zhao,Jing Liu,Lei Zhao,Anquan Ji,Caixia Li. Optimization and validation of analysis method based on 27-plex SNP panel for ancestry inference [J]. Hereditas(Beijing), 2017, 39(2): 166-173. |
| [12] | Feng Gao, Haipeng Li. Application of computer simulators in population genetics [J]. Hereditas(Beijing), 2016, 38(8): 707-717. |
| [13] | Yunsheng Wang. Research progress of plant population genomics based on high-throughput sequencing [J]. Hereditas(Beijing), 2016, 38(8): 688-699. |
| [14] | Xiuyan Ruan, Weini Wang, Yaran Yang, Bingbing Xie, Jing Chen, Yacheng Liu, Jiangwei Yan. Genetic variability and phylogenetic analysis of 39 short tandem repeat loci in Beijing Han population [J]. HEREDITAS(Beijing), 2015, 37(7): 683-691. |
| [15] | WANG Xiao-Qing WANG Chuan-Chao DENG Qiong-Ying LI Hui. Genetic analysis of Y chromosome and mitochondrial DNA poly-morphism of Mulam ethnic group in Guangxi, China [J]. HEREDITAS, 2013, 35(2): 168-174. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||